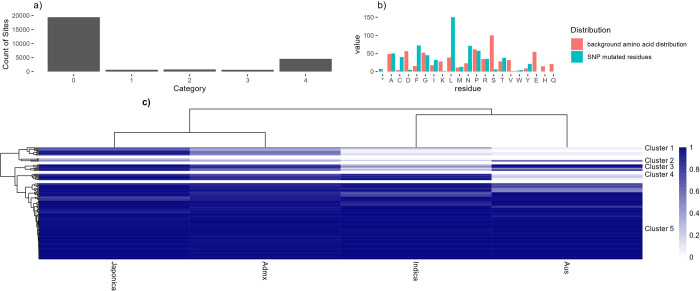
Figure 4.
SAAV analysis. A) Bar chart showing counts of phosphosite by SAAV category. B) Counts of substituted amino acid in category 1 phosphosites, including the normalized background distribution of that amino acid (* = stop codon). C) A heat map to show the assumed major allele frequency (i.e., frequency of Ser/Thr within the 3K “pseudo-proteins” from the rice 3K SNP set) of the phosphosite in four rice family groups: Japonica, Admixed, Indica, and Aus. Allele frequencies are filtered for total difference between families >0.05 to remove genes showing the same frequency across all families.