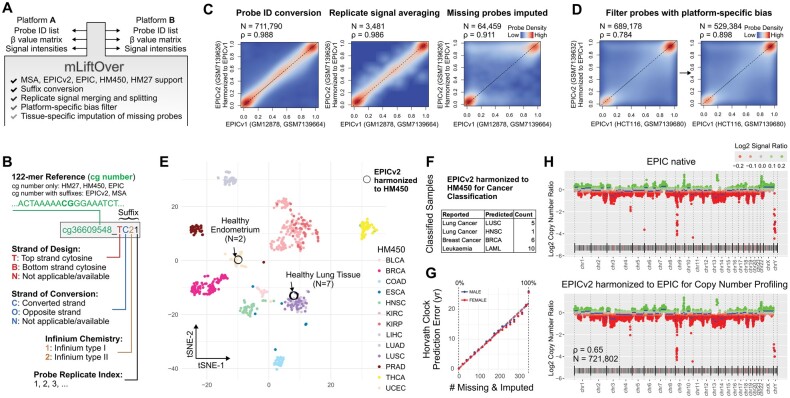
Figure 1.
mLiftOver harmonizes Infinium DNA methylation BeadChip data across array platforms. (A) Schematic illustration of the core features and workflow of mLiftOver from data input to harmonized output. (B) Depiction of the probe naming convention employed in the EPICv2 and MSA arrays. (C) The accuracy of mLiftOver was evaluated using the GM12878 cell line data, contrasting measurements from EPICv1 and EPICv2. The panel is divided into three sub-panels, demonstrating (i) direct probe ID translation, (ii) signal averaging across replicates, and (iii) imputation of missing probe readings (excluding those with methylation level standard deviation >0.08). Spearman’s correlation coefficients are displayed atop each subpanel, with all correlations being significant (P-value <1E-6). (D) Removal of platform-specific biases (tested on a pair of HCT116 cell line data that did not participate in the platform-specific bias analysis), P-value <1E-6. (E) Illustrates the integration process of mLiftOver for primary healthy tissue data and TCGA tumor-adjacent normal tissue data, showcasing its utility in harmonizing diverse datasets for tissue classification. (F) Demonstrates the application of cancer classification models, initially trained on HM450 data using a random forest framework, to primary tumor datasets harmonized from EPICv2 data through mLiftOver. (G) Plot relating the number of missing probes and prediction error of Horvath’s pan-tissue clock, stratified by sex. (H) Compares copy number variation profiles obtained from native EPIC data and profiles harmonized from EPICv2 data, showing the consistency of mLiftOver in signal data conversion.