Abstract
Infertility, a worldwide reproductive health concern, impacts approximately one in five couples. Male infertility, stemming from spermatogenic dysfunction and reduced sperm quality, stands as a primary factor contributing to infertility. Given the global decrease in male fertility linked to environmental factors like the greenhouse effect, it is crucial to develop a comprehensive understanding of how increased temperatures impact both the quantity and quality of sperm. In this study, we utilized Pandora-seq technology to detect the small non-coding RNAs (sncRNAs) expression profile in the testicular tissue of heat-stressed mice. The investigation explores the dynamic shifts in sncRNAs within the mouse testis under heat stress, including miRNAs, tsRNAs, piRNAs, rsRNAs and other sncRNAs. Furthermore, we successfully identified differentially expressed sncRNAs in testicular tissues before and after heat stress. Subsequently, we conducted functional enrichment analysis on the potential predicted target genes of differentially expressed miRNAs and tsRNAs. These datasets will constitute a valuable foundational resource for further investigations into the decline in male reproductive capacity triggered by heat stress.
Subject terms: Non-coding RNAs, Sequencing
Background & Summary
Infertility, namely reproductive failure, has become the main problem of human reproductive health. It is estimated that infertility affects up to one-fifth of couples worldwide1. Among them, infertility caused by male factors accounts for 30–50% of infertility cases2,3. Given the established correlation between semen quality and successful pregnancy, the evaluation of male reproductive capacity often relies on semen quality assessments4–6. Notably, global male fertility is declining rapidly, with average sperm count and concentration dropping by approximately 50%7. Studies have indicated that the greenhouse effect is considered to be the current culprit for the reduction of semen quality in men8,9. Scrotal temperature profoundly impacts male reproductive health and semen quality. Usually, maintaining testicular temperature 2–4 °C lower than the core body temperature is crucial for optimal spermatogenesis. However, when the testis is exposed to heat stress, it accelerates germ cell apoptosis10. The evidence has revealed that for every 1 °C rise in temperature, there’s a correlated decrease in sperm production by 14%, significantly impacting overall sperm count11. Hence, it is imperative to comprehend the underlying causes behind the decline in both quality and quantity of male sperm due to high temperatures. This understanding holds significant importance in averting the decline in male fertility.
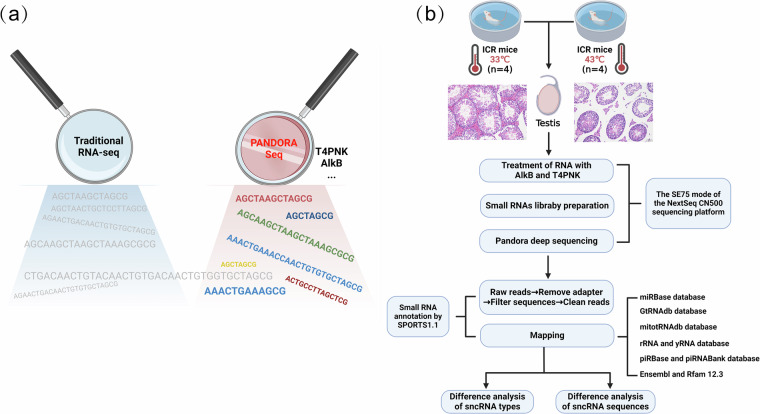
In recent years, there has been a noticeable increase in research attention directed towards understanding the involvement of non-coding RNAs across diverse biological processes12. Non-coding RNAs include long non-coding RNAs (lncRNAs) and small non-coding RNAs (sncRNAs). Over the past decade, high-throughput RNA sequencing technology has significantly advanced the exploration of functional sncRNAs, notably miRNAs. However, when sncRNAs carry specific modifications, including 3′end modification (blocking the adapter ligation process) and RNA methylation modification (interfering with the reverse transcription process), the conversion efficiency of sncRNAs to cDNA is greatly reduced, resulting in reduced sequencing depth and accuracy13,14. To address these challenges, a novel RNA-Seq approach named Pandora-Seq (Panoramic RNA Display by Overcoming RNA modification aborted sequencing) has been developed, effectively circumventing the interference of RNA modifications with sequencing results14. The sequencing approach relies on enzymatic treatment of small RNAs within the 15–50 nucleotide range, employing dealkylase α-ketoglutarate-dependent hydroxylase (AlkB) and its mutant form15, as well as T4 polynucleotide kinase treatment16,17. This process effectively eliminates sequencing-affecting modifications, resulting in a comprehensive profile of sncRNAs (Fig. 1a).
Fig. 1.
Overview of the experimental process. (a) Illustrations of comparison between Pandora-seq and RNA-seq. (b) Illustrations of mouse model construction, sample collection, and Pandora sequencing experimental procedures.
Epigenetic changes, including alterations in sncRNAs, play a pivotal role in gametogenesis and are closely linked to various reproductive disorders18. Recognized as pivotal contributors to epigenetic regulation, sncRNAs have garnered considerable attention for their crucial involvement in testicular development and spermatogenesis19–21. In the context of global warming induced by the greenhouse effect, heat stress factors are impacting male semen quality and fertility8. Hot water bath is a common form of testicular heat stress. Typically, exposure to hot water for over 30 minutes per day can lead to specific damage in the anatomical morphology and sperm quality of the male testis within a week22. Previous studies predominantly delved into specific types of sncRNAs, such as microRNA23 or piRNA24, to understand testicular damage induced by heat stress. However, the dynamic alterations within the intricate regulatory network of sncRNAs in the testis under heat stress remain unexplored. Therefore, to reveal the dynamic changes of the sncRNA library in the mouse testis under heat stress, we collected testicular tissues from both the normal group (NC) and hot water group (HW) mice, obtaining their Pandora sequencing data (Fig. 1b). We analyzed a total of 8 samples, with 4 biological replicates in NC and HW group. Details of the specific samples are presented in Table 1. We used the Illumina NextSeq sequencing platform to generate a total of 278676235 raw reads. The sequencing results were annotated using the small RNA annotation software SPORTS1.1, yielding a total of 99,192,693 match genome reads (Table 2). Our data comprehensively delineate the dynamic changes in small non-coding RNAs (sncRNAs) in testicular tissue before and after heat stress in mice, including miRNAs, tsRNAs, piRNAs, rsRNAs and other sncRNAs.
Table 1.
Basic sample collection information of mice.
| Sample | Age | Weight (g) | Testicular weights (Left, g) | Testicular weights (Right, g) |
|---|---|---|---|---|
| NC-5-T | Ten weeks | 35.93 | 0.144 | 0.136 |
| NC-6-T | Ten weeks | 38.59 | 0.132 | 0.128 |
| NC-8-T | Ten weeks | 33.61 | 0.136 | 0.136 |
| NC-9-T | Ten weeks | 36.51 | 0.134 | 0.145 |
| HW-4-7d-T | Ten weeks | 36.67 | 0.061 | 0.061 |
| HW-5-7d -T | Ten weeks | 37.82 | 0.045 | 0.045 |
| HW-9-7d -T | Ten weeks | 38.36 | 0.065 | 0.065 |
| HW-10-7d -T | Ten weeks | 34.47 | 0.045 | 0.045 |
Table 2.
Summary of PANDORA-Seq Reads.
| Sampe_ID | Raw reads | Clean_Reads | Match_Genome_Reads | miRNA_Match_Reads | rsRNA_Match_Reads | tsRNA_Match_Reads | mt_tsRNA_Match_Reads | piRNA_Match_Reads |
|---|---|---|---|---|---|---|---|---|
| NC-5-T | 21319881 | 12401143 | 8080074 | 55637 | 4703789 | 1294408 | 112163 | 216906 |
| NC-6-T | 23370251 | 13606239 | 8947771 | 57031 | 5307689 | 1356660 | 130939 | 226705 |
| NC-8-T | 23209913 | 13152939 | 8691264 | 56235 | 5036033 | 1371494 | 128175 | 243235 |
| NC-9-T | 25322716 | 13623797 | 8596144 | 29107 | 5478653 | 1150744 | 73845 | 168384 |
| HW-4-7d-T | 21288819 | 11781150 | 7325811 | 90114 | 4958519 | 1172503 | 125238 | 11860 |
| HW-5-7d-T | 20469897 | 11022159 | 7223968 | 84806 | 4851244 | 1079033 | 171800 | 8788 |
| HW-9-7d-T | 25126108 | 13647748 | 8076794 | 111492 | 5333673 | 1042828 | 133340 | 16945 |
| HW-10-7d-T | 24133041 | 13127170 | 8553828 | 83547 | 5390575 | 1068322 | 156340 | 14799 |
Raw reads: Original sequencing readings without quality filtering and clean. Clean reads: SPORTS 1.1 outputs clean reads by removing the sequence adapter, discarding reads obtained by sequences that are longer than the defined range and non-ATUCG sequences. Match Genome Reads: Reads number of raw reads aligned with the reference genome readings. miRNAs match reads: Number of reads aligned to miRNAs sequences. rsRNAs match reads: Number of reads aligned to rRNAs sequences. tsRNAs match reads: Number of reads aligned to mature and precursor tRNAs. mt-tsRNAs match reads: Number of reads aligned to mature and precursor tRNAs in mitochondria. piRNAs match reads: Number of reads aligned to PIWI-interacting RNAs sequences.
Methods
HW mice preparation and sample collection
In this study, we selected 8 ten-week-old male ICR mice (36.50 ± 1.79 g) from DOSSY Experimental Animals Co., Ltd. (Chengdu, China). To establish the testicular heat stress model, mice underwent a daily 25-minute immersion in 43 °C hot water for one week, while the control group experienced baths in 33 °C water. One week following the hot water bath treatment, eight ICR mice were euthanized, and their testicular and epididymal tissues were promptly collected and frozen in liquid nitrogen. The ethical considerations of this study were approved by the Ethics Committee of Sichuan Agricultural University.
Total RNA isolation and quality control
Total RNA was extracted with TRIzol reagent according to manufacturer’s instructions (Invitrogen; 15596018). Agarose gel electrophoresis was utilized to evaluate the quality of the total RNA sample, followed by the isolation of RNA of the specified size from the total RNA. Concurrently, RNA concentration was determined using a Nanodrop spectrophotometer (NanoDrop 2000, Thermo Fisher Science, USA).
Treatment of RNA with AlkB and T4PNK
Before constructing the small RNA-seq library and performing deep sequencing, the purified small RNA was treated with T4PNK and AlkB to address the issue of RNA modification hindering the passability of reverse transcriptase. Initially, RNA underwent treatment with AlkB enzyme. The RNA was incubated in 50 μL reaction mixture containing 50 mM HEPES (pH 8.0) (Gibco (15630080) and Alfa Aesar (J63578), 75 μM ferrous ammonium sulfate (pH 5.0), 1 mM α-ketoglutaric acid (Sigma-Aldrich; K1128-25G), 2 mM sodium ascorbate, 50 mg l−1 bovine serum albumin (Sigma-Aldrich; A7906-500G), 4 μg ml−1 AlkB (Guangzhou Epibiotek Co., Ltd., Guangzhou, China), 2,000 U RNase inhibitor and 200 ng RNA at 37 °C for 30 min. Then, the mixture was added into 500 μL TRIzol reagent to perform the RNA isolation procedure. Subsequently, RNA was subjected to treatment with T4PNK. The RNA was incubated in 50 μL reaction mixture containing 5 μL 10 × PNK buffer (New England Biolabs; B0201S), 10 mM ATP (New England Biolabs; P0756S), 10 U T4PNK (New England Biolabs; M0201L) and 200 ng RNA at 37 °C for 20 min. Then, the mixture was added into 500 μL TRIzol reagent to perform the RNA isolation procedure.
Small RNA library construction and deep sequencin
The RNA segment was separated by PAGE, then a 15- to 45-nucleotide stripe was selected and recycled. The adapters were obtained from the QIAseq® miRNA Library Kit (QIAGEN: 331505) and ligated sequentially. The amplified flow cell was sequenced on the Illumina system by epibiotek (Guangzhou Epibiotek Co., Ltd., Guangzhou, China). The raw sequencing data were generated using the SE75 mode of the NextSeq CN500 sequencing platform.
Small RNA annotation and analyses for PANDORA-seq data
The annotation of small RNA sequences was carried out using the non-coding RNA annotation and analysis software SPORTS1.1, specifically optimized for small RNAs derived from rRNA and tRNA. Reads were mapped to the following individual non-coding RNA databases sequentially: (1) the microRNA database miRBase 21; (2) the rRNA and YRNA databases assembled from the National Center for Biotechnology Information nucleotide and gene database; (3) the genomic tRNA database GtRNAdb; (4) the mitochondrial tRNA database mitotRNAdb; (5) the piRNA database, including piRBase and piRNABank; (6) the non-coding RNAs defined by Ensembl and Rfam 12.3. In the annotation of tsRNAs, both pre-tRNA and mature tRNA sequences were employed for comprehensive annotation. Mature tRNA sequences were derived from the GtRNAdb and mitotRNAdb sequences using the following procedures: (1) predicted introns were removed; (2) a CCA sequence was added to the 3′ ends of all tRNAs; and (3) a G nucleotide was added to the 5′ end of histidine tRNAs. The tsRNAs were categorized into four types based on the origin of the tRNA loci: (1) 5′ tsRNA (derived from the 5′ end of pre-/mature tRNA); (2) 3′ tsRNA (derived from the 3′ end of pre-tRNA); (3) 3′ tsRNA-CCA end (derived from the 3′ end of mature tRNA); (4) and internal tsRNAs (not derived from 3′ or 5′ loci of tRNA). For rsRNA annotation, rsRNAs derived from 4.5 S, 5 S, 5.8 S, 12 S, 16 S, 18 S, 28 S, and 45 S rRNA were distinguished based on the sequence fragment size of rRNA. Mapping to the parent rRNA in ascending order of rRNA sequence length will substantially ensure the uniqueness of each rsRNA annotation. For tsRNA annotation, annotation was based on pre-tRNA and mature tRNA. DESeq 2 was employed to identify sncRNAs with significant differences, using screening criteria of log2(Fold Change) > 1 and P < 0.05.
Dynamic changes of sncRNAs expression and sequence characteristics in mouse testicular tissue before and after heat stress
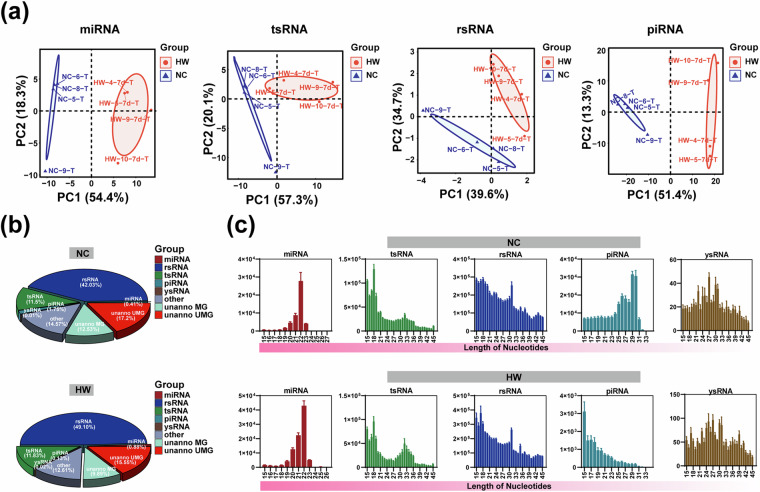
Initially, we performed principal component analysis (PCA) on the read counts of miRNA, tsRNA, rsRNA, and piRNA for each sample (Fig. 2a). The PCA results highlighted significant differences in the distribution of testicular tissue samples before and after heat stress across each sncRNA family. By analyzing the read counts of diverse sncRNAs, we characterized the expression profile of sncRNAs in mouse testicular tissue before and after exposure to heat stress. The pie chart illustrates the proportion distribution of different types of sncRNAs in testicular tissue before and after heat stress treatment (Fig. 2b). The results revealed that upon the onset of testicular heat stress, the proportion of rsRNA expression increased from 42.03% to 49.10%, whereas the proportion of piRNA expression dropped from 1.75% to 0.13%. We further investigated the nucleic acid length distribution for each subtype of sncRNAs (Fig. 2c). Specifically, miRNA lengths were concentrated around 22 nt, tsRNA exhibited distinct peaks at 18 nt and 32 nt, rsRNA displayed a peak at 31 nt, and ysRNA presented a peak with relatively less clarity. Interestingly, the peak value of piRNA was evident at 29 nt or 30 nt before the induction of heat stress. However, upon the occurrence of heat stress, there was a noticeable reduction in the overall expression of piRNA, and the peak value became less defined.
Fig. 2.
The expression and sequence characteristics of sncRNAs in testicular tissue of mice before and after heat stress. (a) Principal component analysis (PCA) based on the reading counts of various sncRNAs in the sample. (b) The distribution of sncRNAs types in samples before and after heat stress was expressed in proportion. (c) Length distribution of various types of sncRNAs in samples before and after heat stress.
The sncRNA landscape of rsRNA and tsRNA before and after heat stress in mice
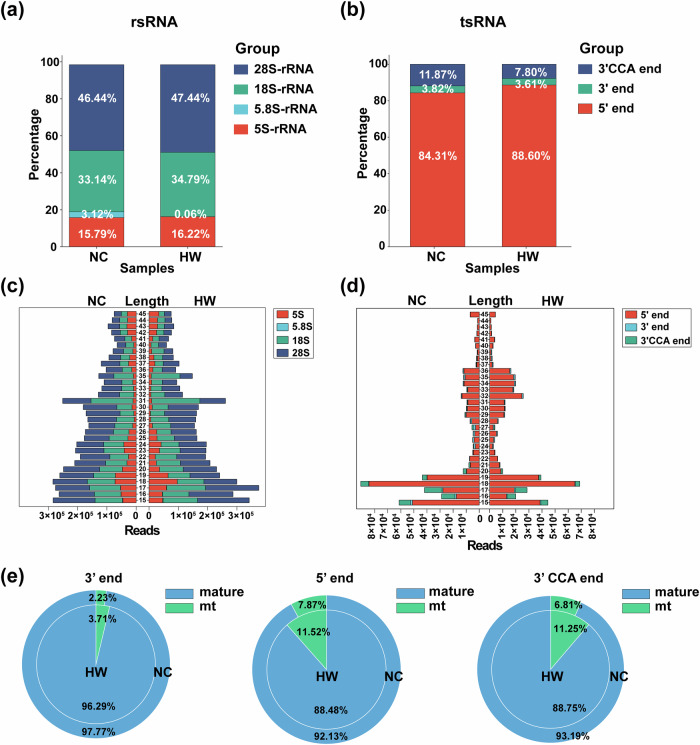
Taking into consideration the diverse types of rsRNA and tsRNA, we conducted a detailed analysis of the subtypes within these two categories of sncRNAs. Figure 3a illustrated the distribution of the number of rsRNAs derived from fragments of different lengths. The absolute proportion of rsRNA originating from 28 S rRNA was approximately 50% in both sample groups. Furthermore, following heat stress in mouse testis, there was a notable decrease in the proportion of rsRNA derived from 5.8 S rRNA. It is worth noting that, as previously illustrated in Fig. 2b, the proportion of rsRNA expression increased under heat stress conditions. Similarly, we analyzed the distribution of various types of tRNA derived fragments in the samples (Fig. 3b). The results indicated that heat stress treatment elevated the proportion of 5′ tsRNA in mouse testicular tissue while decreasing the proportion of 3′ CCA tsRNA. Additionally, we visualized the sequence length distribution for each type of rsRNA and tsRNA (Fig. 3c,d). We also conducted an analysis of the sources of different types of tsRNA, which include pre-tRNA, mature tRNA, and mitochondrial tRNA. Due to the exceptionally low content of pre-tRNA, it is not represented in the pie chart (Fig. 3e). Interestingly, heat stress appears to have no influence on the content of 3′ tsRNA from diverse sources in the mouse testis. However, heat stress led to a reduction in the expression of 5′ tsRNA and 3′ CCA tsRNA derived from mature tRNA, while simultaneously increasing the expression of 5′ tsRNA and 3′ CCA tsRNA derived from mitochondrial tRNA.
Fig. 3.
The sncRNA landscape of rsRNA and tsRNA before and after heat stress in mice. (a) Expression patterns of rsRNA fragments in samples before and after heat stress. (b) Expression patterns of tsRNA types in samples before and after heat stress. (c) Distribution of sequence lengths among rsRNA fragment subtypes. (d) Distribution of sequence lengths for tsRNA types. (e) Distribution of tsRNA type sources.
Identification of differentially expressed sncRNAs
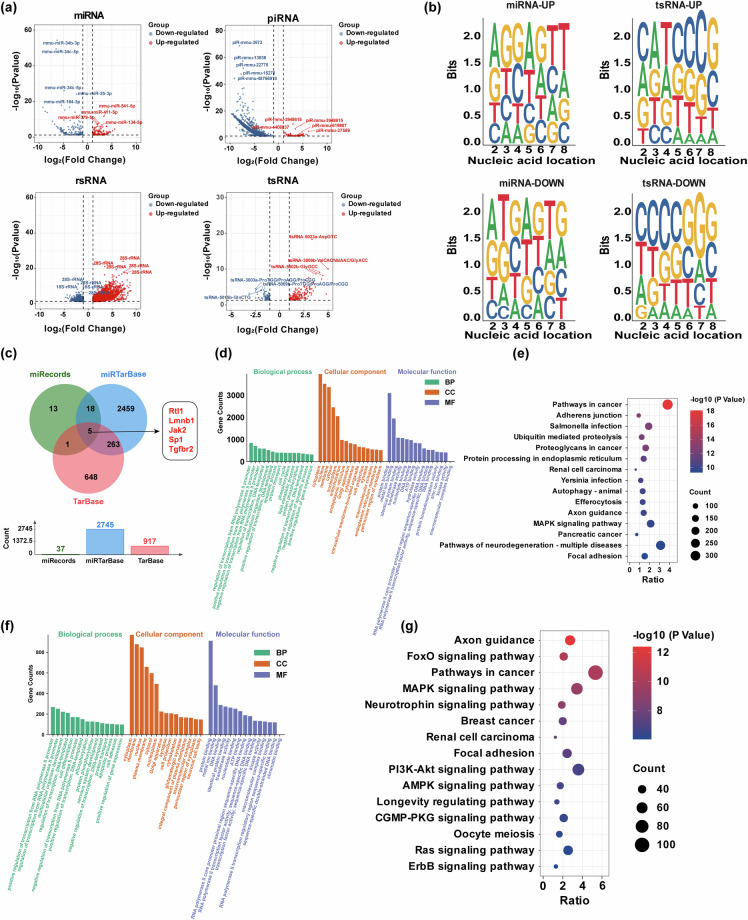
Differentially expressed sncRNA analysis was performed using the R package DESeq 2. The criteria used for screening differentially expressed sncRNAs were log2(Fold Change) > 1 and P value < 0.05. We visualized the differentially expressed sncRNAs based on the screening conditions. The red circle represents up-regulated sncRNAs in the HW group, while the blue circle represents down-regulated sncRNAs (Fig. 4a). Simultaneously, utilizing P values as a criterion, we identified and labeled the top five up-regulated and down-regulated sncRNAs with the most significant differences. Given that miRNA and tsRNA exert their functions relying on the 5′-end seed sequence, we conducted an analysis of the base preference of the seed sequence in miRNA and tsRNA (Fig. 4b). We observed a significant difference in the base site preference of seed sequences for differentially expressed miRNAs and tsRNAs between the two sample groups before and after heat stress. For differentially expressed miRNAs (Threshold: log2FC > 1.5 and P value < 0.05), we predicted their potential target genes using three databases: miRecords, miRTarBase, and TarBase (https://jingege.shinyapps.io/jingle_molecular/). A total of 3408 validated target genes were predicted, and five of these target genes were consistently displayed in all three databases (Fig. 4c). For potential target genes of miRNAs, we conducted GO and KEGG functional enrichment analysis using the DAVID website (https://david.ncifcrf.gov/summary.jsp). GO functional enrichment analysis revealed that the potential target genes of miRNAs were predominantly enriched in biological processes such as transcriptional regulation, cell survival and death, and protein function and transport (Fig. 4d). KEGG analysis demonstrated that the potential target genes of differentially expressed miRNAs, both before and after heat stress, were predominantly enriched in pathways related to signal transduction, cell survival, and metabolism (Fig. 4e). Similarly, for differentially expressed tsRNAs, we predicted their potential target genes using the online prediction website TargetScan (https://www.targetscan.org/vert_50/seedmatch.html) and conducted GO and KEGG functional enrichment analysis using the DAVID website (https://david.ncifcrf.gov/summary.jsp). GO functional enrichment analysis revealed that the potential target genes of tsRNA were predominantly enriched in biological processes such as transcriptional regulation, cell development and differentiation, and cell survival and death (Fig. 4f). KEGG analysis showed that the potential target genes of differentially expressed tsrna before and after heat stress were mainly enriched in pathways related to signal transduction, cytoskeleton and signal regulation, and cell metabolism (Fig. 4g).
Fig. 4.
Conserved tsRNAs in adipose tissue of pigs and mice. (a) Volcano plot of differentially expressed sncRNAs between NC and HW groups. (b) Bias of differentially expressed miRNA and tsRNA seed sequences. (c) Venn diagram of potential target genes predicted by differentially expressed miRNAs in three databases. (d) GO enrichment analysis of potential target genes of miRNAs. Each type showed the enrichment results of the top 15 of gene count. (e) KEGG analysis of potential target genes of miRNAs. Only the top 15 signaling pathways with the most significant differences were showed. (f) GO enrichment analysis of potential target genes of tsRNAs. Each type showed the enrichment results of the top 15 of gene count. (g) KEGG analysis of potential target genes of tsRNAs. Only the top 15 signaling pathways with the most significant differences were showed.
Data Records
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in National Genomics Data Center (Nucleic Acids Res 2022), China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA014959) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa25.
Technical Validation
RNA quality control
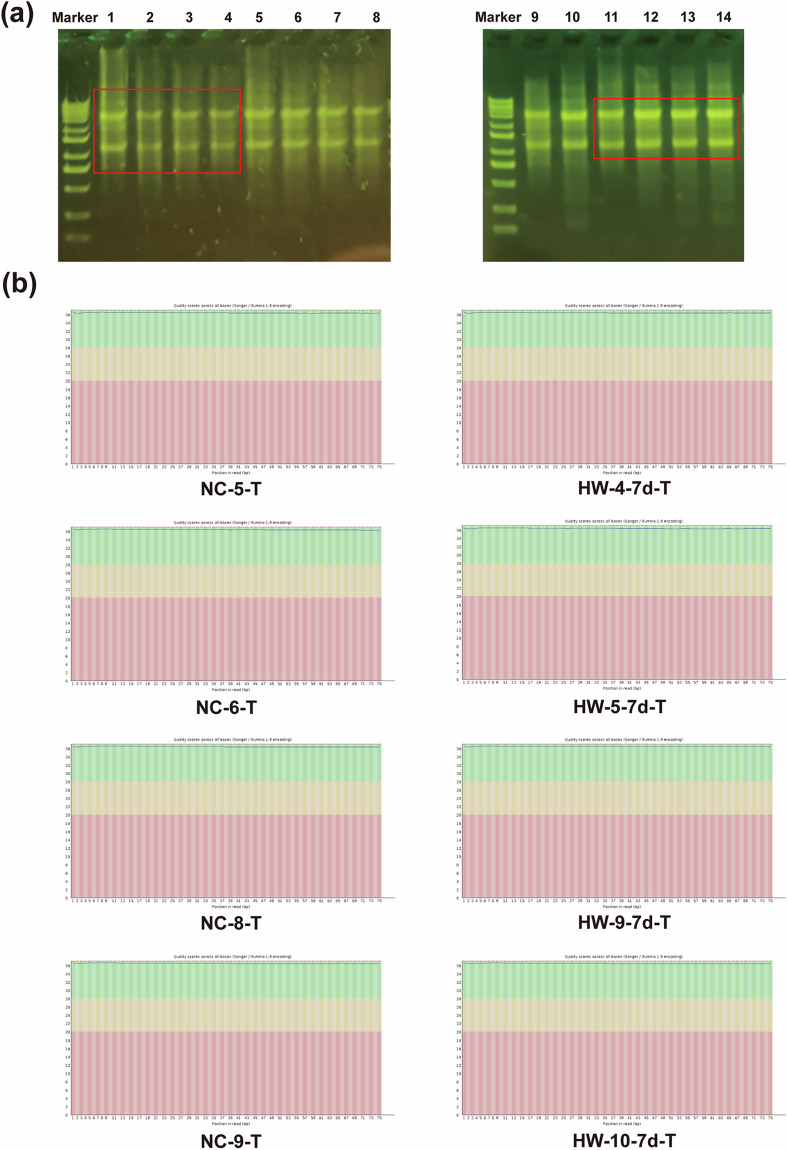
The purity and size of the extracted RNA were assessed using agarose gel electrophoresis to ensure the integrity of the samples utilized for constructing both the small RNA library and Pandora sequencing. The detailed information for each RNA sample was presented in Table 3, and the original agarose gel electrophoresis results were provided in Fig. 5a. The results indicated the good quality of our RNA samples, and the electrophoresis conditions were deemed suitable, ensuring the reliability of subsequent Pandora sequencing samples.
Table 3.
Sample RNA quality test results.
| Sample | Concentration (ng/μL) | Volume (μL) | Total volume (ng) | Quality inspection results |
|---|---|---|---|---|
| NC-5-T | 8160 | 40 | 326400 | Qualified |
| NC-6-T | 7320 | 40 | 292800 | Qualified |
| NC-8-T | 7320 | 40 | 292800 | Qualified |
| NC-9-T | 7560 | 40 | 302400 | Qualified |
| HW-4-7d-T | 4260 | 40 | 170400 | Qualified |
| HW-5-7d-T | 3600 | 40 | 144000 | Qualified |
| HW-9-7d-T | 1584 | 40 | 63360 | Qualified |
| HW-10-7d-T | 2790 | 40 | 111600 | Qualified |
Fig. 5.
RNA agarose gel electrophoresis results and Pandora sequencing data quality control chart. (a) Agarose gel electrophoresis: Lanes 1–4 represent samples NC-5-T, NC-6-T, NC-8-T, NC-9-T, and lanes 11–14 represent samples HW-5-7d-T, HW-4-7d-T, HW-9-7d-T, HW-10-7d-T, respectively. (b) The X-axis represents the sequencing position of the base, and the Y-axis represents the sequencing quality score. The yellow area signifies a 1% error reading rate, and the green area represents a 0.1% error reading rate, indicating high data quality.
Sequencing quality control
The raw sequencing data in FastQC format were obtained from the Illumina sequencing platform. Quality assessment of the sequencing data was performed using FastQC software (version 0.11.8). We created a per-base sequence quality distribution map for each sample. The y-axis represents the sequencing quality score (expressed as Q), with Q = 20% indicating an error read rate of 1%, and Q = 30% indicating an error read rate of 0.1%. The results indicated that Q > 30%, signifying good quality of the sequencing data (Fig. 5b).
Reproducibility Validation
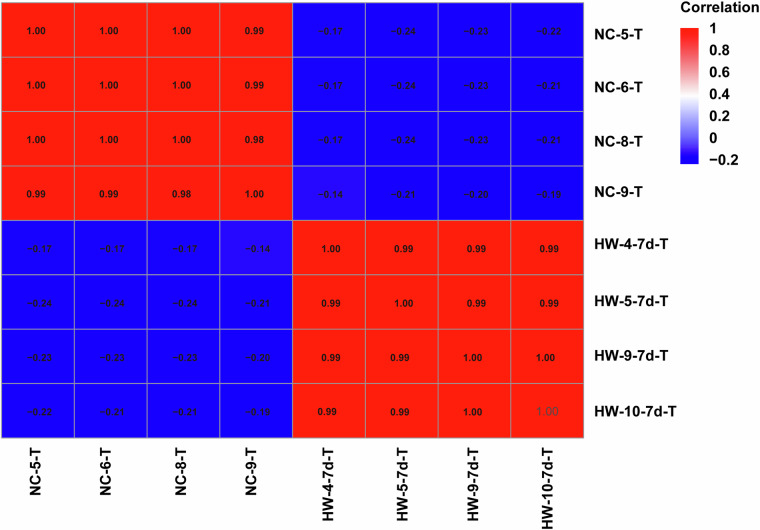
To verify the biological repeatability of mouse testicular tissue samples before and after heat stress, we performed Pearson correlation coefficient analysis on all 8 samples. The correlation heatmap indicated a high correlation coefficient among biological replicates for both before and after heat stress samples (Fig. 6). Furthermore, principal component analysis (PCA) demonstrated distinct separation between samples before and after heat stress, with biological samples from the same treatment forming clearly clustered groups (Fig. 2a). These results affirm the high confidence and reliability of our research data.
Fig. 6.
Correlation heat map of mouse testicular samples before and after heat stress.
Acknowledgements
This work was supported by National Natural Science Foundation of China (32302689); Sichuan Science and Technology Program (2021YFYZ0030, 2021YFYZ0007, 2024NSFSC1176); the Program for Pig Industry Technology System Innovation Team of Sichuan Province (sccxtd-2024-08-09); China Agriculture Research System (CARS-35).
Author contributions
M.G. and Y.L. designed the experiment framework and completed data collation, visual analysis, and manuscript writing. K.W. constructed a mouse testicular heat stress model, collected samples, and established a sequencing library. Y.W. participated in data review and formal analysis (biostatistics). T.L. and J.M. assisted in the conception, drafting and revision of manuscripts. L.Z. and L.S. participated in experimental design, supervised manuscript writing, project management, review and proofreading, and fund acquisition.
Code availability
In this study, open-source bioinformatics analysis software was employed to analyze the raw sequencing data. Additionally, custom scripts or code were not utilized for dataset management and validation. Details of all software toolkits used are as follows:
FastQC (version 0.11.8) was utilized for assessing the quality of the raw sequencing data (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
Bioinformatics provides an online analysis tool for principal component analysis (PCA), correlation analysis, pie charts, histogram pyramid stacks, and other functionalities (www.bioinformatics.com).
The seed sequence motif was done using the Hiplot (ORG) online tool (https://hiplot.cn/).
Online prediction websites, including miRecords26, miRTarBase27, and TarBase28 were employed to predict potential target genes of miRNAs.
The potential target gene prediction of tsRNA was performed on the online prediction website Targetscan (https://www.targetscan.org/vert_50/seedmatch.html).
DAVID online analysis website was employed for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analysis (https://david.ncifcrf.gov/summary.jsp).
R software was used for differential expression analysis of differentially expressed sncRNAs (https://www.r-project.org/).
GraphPad Prism (version 8.0.2, GraphPad Software Inc., USA) was used for statistical analyses and data visualization.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Mailin Gan, Yuhang Lei, Kai Wang.
Contributor Information
Li Zhu, Email: zhuli@sicau.edu.cn.
Linyuan Shen, Email: shenlinyuan@sicau.edu.cn.
References
- 1.WHO TEAM: Sexual and Reproductive Health and Research. Infertility Prevalence Estimates, 1990–2021 (WHO, 2023).
- 2.Punab M, et al. Causes of male infertility: a 9-year prospective monocentre study on 1737 patients with reduced total sperm counts. Hum Reprod. 2017;32:18–31. doi: 10.1093/humrep/dew284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agarwal A, Mulgund A, Hamada A, Chyatte MR. A unique view on male infertility around the globe. Reprod Biol Endocrinol. 2015;13:37. doi: 10.1186/s12958-015-0032-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guzick DS, et al. Sperm morphology, motility, and concentration in fertile and infertile men. N Engl J Med. 2001;345:1388–1393. doi: 10.1056/NEJMoa003005. [DOI] [PubMed] [Google Scholar]
- 5.Small DR, Collins JA, Wilson EH, Wrixon W. Interpretation of semen analysis among infertile couples. CMAJ. 1987;136:829–833. [PMC free article] [PubMed] [Google Scholar]
- 6.Sripada S, et al. Relationship between semen parameters and spontaneous pregnancy. Fertil Steril. 2010;94:624–630. doi: 10.1016/j.fertnstert.2009.02.085. [DOI] [PubMed] [Google Scholar]
- 7.Levine H, et al. Temporal trends in sperm count: a systematic review and meta-regression analysis of samples collected globally in the 20th and 21st centuries. Hum Reprod Update. 2023;29:157–176. doi: 10.1093/humupd/dmac035. [DOI] [PubMed] [Google Scholar]
- 8.Zhou Y, et al. Association between ambient temperature and semen quality: A longitudinal study of 10 802 men in China. Environ Int. 2020;135:105364. doi: 10.1016/j.envint.2019.105364. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X, et al. Association between ambient temperature and semen quality among sperm donation volunteers in South China. Environ Int. 2023;173:107809. doi: 10.1016/j.envint.2023.107809. [DOI] [PubMed] [Google Scholar]
- 10.Yin Y, Hawkins KL, DeWolf WC, Morgentaler A. Heat stress causes testicular germ cell apoptosis in adult mice. J Androl. 1997;18:159–165. doi: 10.1002/j.1939-4640.1997.tb01896.x. [DOI] [PubMed] [Google Scholar]
- 11.Wang C, et al. Effect of increased scrotal temperature on sperm production in normal men. Fertil Steril. 1997;68:334–339. doi: 10.1016/S0015-0282(97)81525-7. [DOI] [PubMed] [Google Scholar]
- 12.Kugel JF, Goodrich JA. Non-coding RNAs: key regulators of mammalian transcription. Trends Biochem Sci. 2012;37:144–151. doi: 10.1016/j.tibs.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai Q, Zheng G, Schwartz MH, Clark WC, Pan T. Selective Enzymatic Demethylation of N2,N2 -Dimethylguanosine in RNA and Its Application in High-Throughput tRNA Sequencing. Angew Chem Int Ed Engl. 2017;56:5017–5020. doi: 10.1002/anie.201700537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi J, et al. PANDORA-seq expands the repertoire of regulatory small RNAs by overcoming RNA modifications. Nat Cell Biol. 2021;23:424–436. doi: 10.1038/s41556-021-00652-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cozen AE, et al. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat Methods. 2015;12:879–884. doi: 10.1038/nmeth.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akat, K. M. et al. Detection of circulating extracellular mRNAs by modified small-RNA-sequencing analysis. JCI Insight5, e127317, 127317 (2019). [DOI] [PMC free article] [PubMed]
- 17.Giraldez MD, et al. Phospho-RNA-seq: a modified small RNA-seq method that reveals circulating mRNA and lncRNA fragments as potential biomarkers in human plasma. EMBO J. 2019;38:e101695. doi: 10.15252/embj.2019101695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bourc’his D, Voinnet O. A small-RNA perspective on gametogenesis, fertilization, and early zygotic development. Science. 2010;330:617–622. doi: 10.1126/science.1194776. [DOI] [PubMed] [Google Scholar]
- 19.Sellem E, et al. Dynamics of cattle sperm sncRNAs during maturation, from testis to ejaculated sperm. Epigenetics Chromatin. 2021;14:24. doi: 10.1186/s13072-021-00397-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.La Y, et al. Identification and Characterization of Piwi-Interacting RNAs for Early Testicular Development in Yak. Int J Mol Sci. 2022;23:12320. doi: 10.3390/ijms232012320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McIver SC, Roman SD, Nixon B, McLaughlin EA. miRNA and mammalian male germ cells. Hum Reprod Update. 2012;18:44–59. doi: 10.1093/humupd/dmr041. [DOI] [PubMed] [Google Scholar]
- 22.Kanter M, Aktas C, Erboga M. Heat stress decreases testicular germ cell proliferation and increases apoptosis in short term: an immunohistochemical and ultrastructural study. Toxicol Ind Health. 2013;29:99–113. doi: 10.1177/0748233711425082. [DOI] [PubMed] [Google Scholar]
- 23.Gan M, et al. Potential Function of Testicular MicroRNAs in Heat-Stress-Induced Spermatogenesis Disorders. Int J Mol Sci. 2023;24:8809. doi: 10.3390/ijms24108809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen W, et al. Exposure to elevated temperature affects the expression of PIWI-interacting RNAs and associated transcripts in mouse testes. Andrology. 2023;11:724–737. doi: 10.1111/andr.13381. [DOI] [PubMed] [Google Scholar]
- 25.2024. Genome Sequence Archive. https://ngdc.cncb.ac.cn/gsa/browse/CRA014959
- 26.Xiao F, et al. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37:D105–110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu S-D, et al. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011;39:D163–169. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vlachos IS, et al. DIANA-TarBase v7.0: indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res. 2015;43:D153–159. doi: 10.1093/nar/gku1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- 2024. Genome Sequence Archive. https://ngdc.cncb.ac.cn/gsa/browse/CRA014959
Data Availability Statement
In this study, open-source bioinformatics analysis software was employed to analyze the raw sequencing data. Additionally, custom scripts or code were not utilized for dataset management and validation. Details of all software toolkits used are as follows:
FastQC (version 0.11.8) was utilized for assessing the quality of the raw sequencing data (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
Bioinformatics provides an online analysis tool for principal component analysis (PCA), correlation analysis, pie charts, histogram pyramid stacks, and other functionalities (www.bioinformatics.com).
The seed sequence motif was done using the Hiplot (ORG) online tool (https://hiplot.cn/).
Online prediction websites, including miRecords26, miRTarBase27, and TarBase28 were employed to predict potential target genes of miRNAs.
The potential target gene prediction of tsRNA was performed on the online prediction website Targetscan (https://www.targetscan.org/vert_50/seedmatch.html).
DAVID online analysis website was employed for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analysis (https://david.ncifcrf.gov/summary.jsp).
R software was used for differential expression analysis of differentially expressed sncRNAs (https://www.r-project.org/).
GraphPad Prism (version 8.0.2, GraphPad Software Inc., USA) was used for statistical analyses and data visualization.