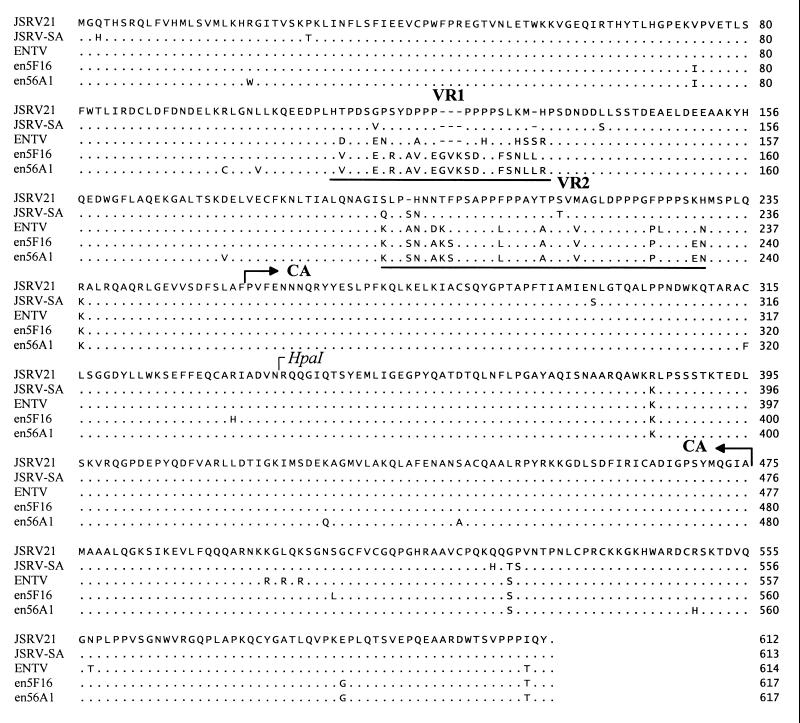
FIG. 2.
Alignment of deduced gag amino acid sequences of sheep type D retroviruses: exogenous JSRV21 (AF105220), JSRV-SA (M80216), and ENTV (Y16627) and the endogenous enJS5F16 and enJS56A1 that maintain full-length gag open reading frames. Dots refer to identical sequences, while dashes indicate lack of sequence. VR1 and VR2 are underlined. Note that the prolines in VR1 of the exogenous JSRVs and ENTV are absent in the endogenous proviruses. In VR2, ENTV is more similar to enJSRVs than JSRV. The putative CA region and the HpaI site used to generate exogenous-endogenous chimeras (Fig. 4) are indicated.