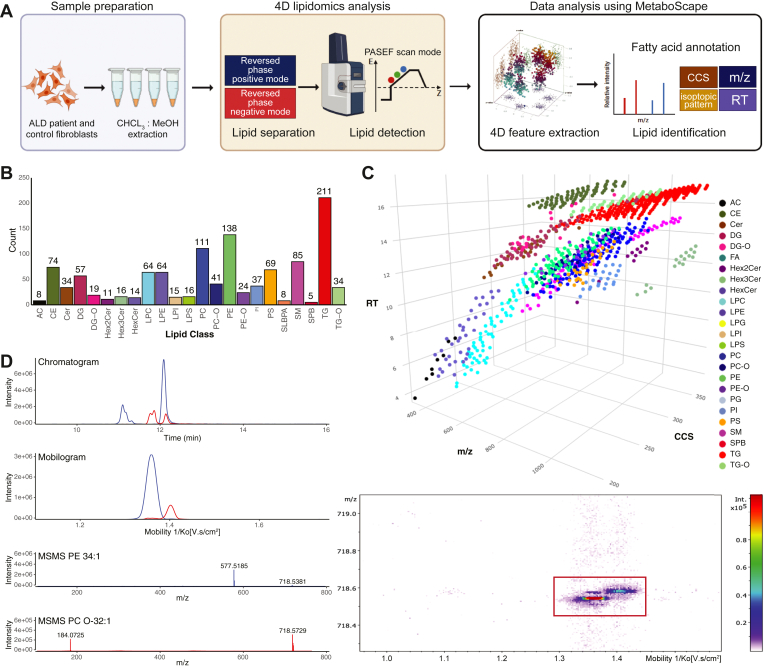
Fig. 1.
4D lipidomics profiling using PASEF (A) 4D lipidomics workflow. Samples are extracted using methanol:chloroform, followed by LC and TIMS mass spectrometry. Lipids are detected using PASEF which enables efficient precursor selection and MSMS acquisition. Features are identified using accurate mass, retention time, isotopic pattern, MSMS spectra, and CCS. B: Bar chart of detected lipid species per class. C: Three-dimensional representation (RT, m/z, CCS) of detected lipids. Different colors represent different lipid classes. D: Chromatogram of PE 34:1 (blue) and PC O-32:1 (red) show overlapping peaks in the LC dimension but are not overlapping in the ion mobility dimension. PASEF enables MSMS coverage for nearly every ion mobility resolved peak leading to noninterfering MSMS spectra of PE 34:1 and PC O-32:1. CCS, collisional cross section; PASEF, parallel accumulation serial fragmentation; PC, phosphatidylcholine; PE, phosphatidylethanolamine; RT, retention time; TIMS, trapped ion mobility spectrometry.