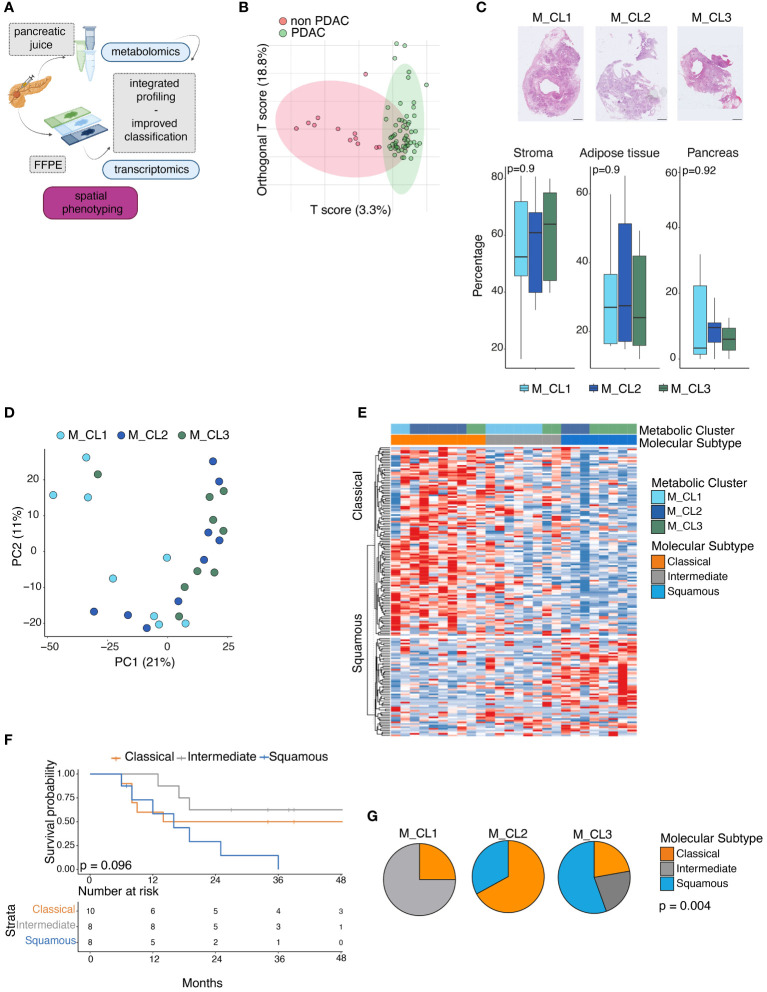
Figure 1.
Metabolic and transcriptomic analysis of PDAC paired biospecimens. (A) Schematic of the work. Transcriptomic analysis of 26 PDAC sections, on whose pancreatic juice metabolomic analysis had been performed. Integration of multiomics information can improve patient profiling. (performed with Biorender.com) (B) OPLS-DA score plot obtained from the metabolic profiling of 70 pancreatic juices (12 non PDAC, 58 PDAC). (C) Percentage of stromal tissue, adipose tissue and pancreatic glands at the surgical resection margin in the three metabolic groups. Representative pictures of Hematoxylin and Eosin (H&E) staining on tissue slides from M_CL1, M_CL2, M_CL3 residual pancreatic tissue (up). Scale bar: 2mm. Quantification of the three tissue regions by imaging analysis in the three groups (bottom). P=ns by t test. (D) Principal component analysis projection of 26 PDAC samples colored by Metabolic cluster assignment. (E) Gene set variation analysis (GSVA) of classical and squamous genes, in 26 PDAC samples, according to the Moffitt molecular subtyping. Heatmap showing enrichment scores across 26 samples, color-coded by metabolic cluster and molecular subtype. Samples with score difference lower than 3 are classified as intermediate. (F) Survival analysis of patients classified according to the molecular profile. P=0.096 by log-rank test. (G) Pie charts showing proportions of molecular subtypes (classical, intermediate and squamous) in the metabolic groups. p= 0.004 by Kruskal-Wallis rank sum test.