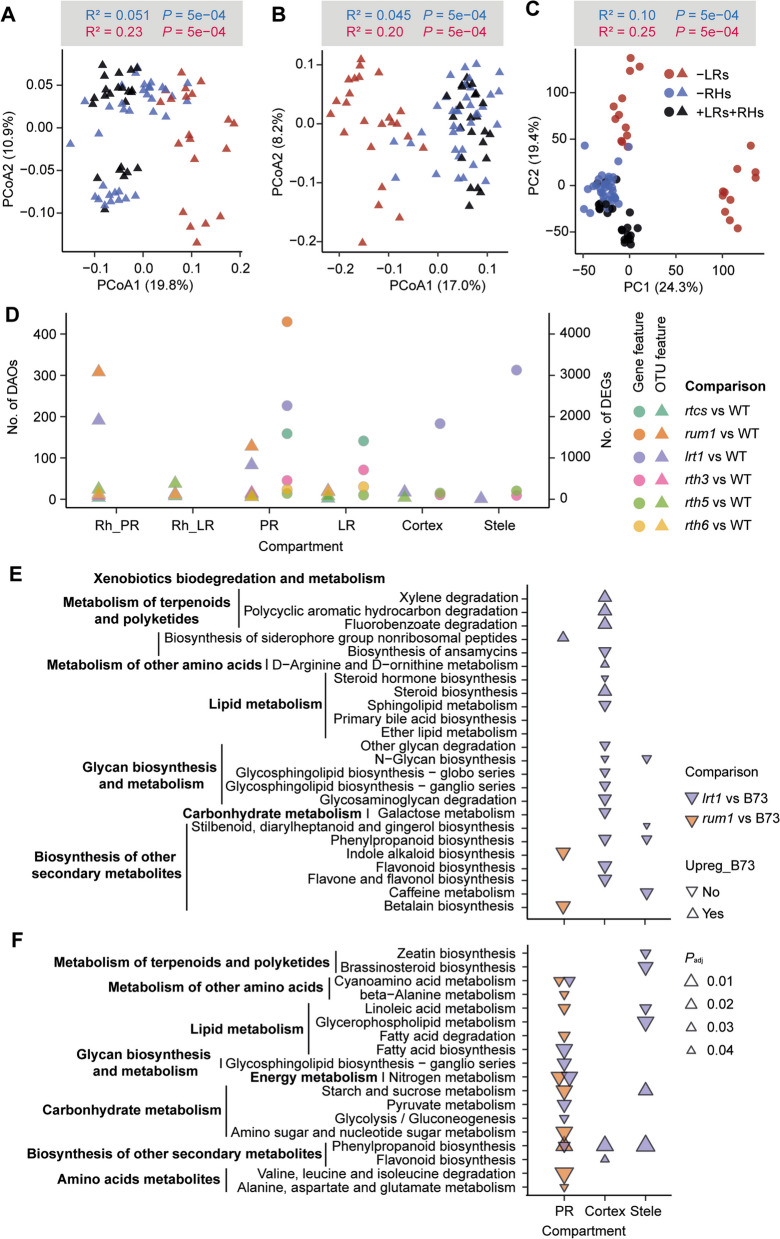
Fig. 2.
Lateral roots determine microbiome assemblage and transcriptomic changes across different compartments. Principal coordinate analysis (PCoA) showing the effects of root hairs and lateral root defects on the dissimilarity of bacterial β-diversity across 3 groups (WT: n = 23; Lateral root mutants: n = 20; Root hair mutants: n = 35) in the rhizosphere (A) and 3 groups (WT: n = 24; Lateral root mutants: n = 24; Root hair mutants: n = 36) in the root (B) compartments from primary root. C Principal component analysis (PCA) illustrating the effects of root hairs and lateral root defects on transcriptomic shift across 3 groups (WT: n = 24; lateral root mutants: n = 23; root hair mutants: n = 33) along the primary root. For both, the bacterial and transcriptomic dissimilarity matrix data, the explained variance by the effects from root hair and lateral roots were assessed by pair-wise permutational analysis of variance (PERMANOVA, P < 0.001). D Pair-wise comparison of differentially abundant OTUs (DAOs) and differentially expressed genes (DEGs) between each monogenic mutant and wild type (WT). The triangles and dots indicated the microbiome and transcriptome features respectively. Rh_PR, rhizosphere from the primary root; Rh_LR, rhizosphere from lateral root; PR, primary root; LR, lateral root. OTU, operational taxonomic unit. E Prediction of the functional potential of the bacterial microbiota using the PICRUSt tool from the lateral root mutants in comparison to the wild-type plants. Directions of triangles indicate up- and downregulation in mutant and wild-type B73. F Prediction of root metabolism using the KEGG pathway analysis from the lateral root mutants in comparison to the wild-type plant. The size of the triangles indicates the significance degree. The purple and orange color indicates the lrt1 and rum1 compared with wild-type B73 respectively