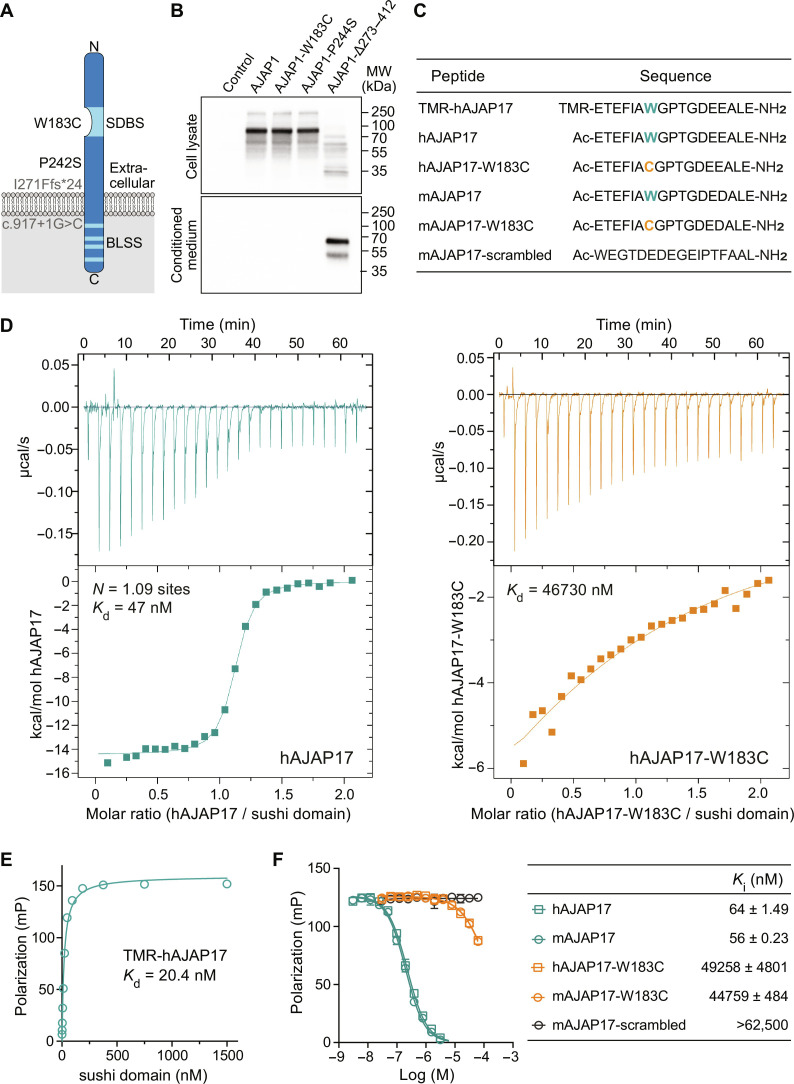
Fig. 1. In vitro characterization of AJAP1 variants.
(A) Scheme depicting patient AJAP1 variants. The W183C variant is located in the SDBS. The P242S variant, corresponding to mouse AJAP1-P244S, affects a residue in the extracellular domain. Hypothetical proteins are depicted in gray. The possible protein product of the p.I271Ffs*24 variant, corresponding to mouse AJAP1-Δ273–412, is hypothesized to generate a protein lacking both the transmembrane domain and the intracellular domain containing BLSS. The AJAP1 splice variant c.917+1G>C is hypothesized to generate a protein lacking the intracellular domain after Cys306. However, both I271Ffs*24 and c.917+1G>C may also trigger nonsense-mediated decay. (B) Immunoblots of mouse AJAP1 variants expressed under identical conditions in HEK293T cells. Top: Cell lysates of AJAP1-, AJAP1-W183C–, and AJAP1-P244S–expressing cells. Bottom: Secreted AJAP1-Δ273–412 protein in conditioned cell culture medium. AJAP1 proteins were detected using a polyclonal anti-AJAP1 antibody (AF7970, R&D Systems). (C) Human (h) and mouse (m) AJAP17 and AJAP17-W183C peptides used in binding experiments. The W183C variant is highlighted in color. TMR-hAJAP17 is N-terminally labeled with the fluorophore Tamra; mAJAP17-scrambled served as a negative control in binding experiments. (D) Representative ITC diagrams of sushi domain protein (9) in solution with increasing amounts of hAJAP17 (blue) or hAJAP17-W183C (orange) peptides. Raw heat signatures (top) and integrated molar heat release (bottom) are shown. The calculated stoichiometry (N) and the Kd are indicated. (E) Saturation binding of sushi domain protein to TMR-hAJAP17 (25 nM) determined by FP analysis. (F) Competition of TMR-hAJAP17 (25 nM) at sushi domain protein (40 nM) by increasing concentrations of unlabeled peptides determined by FP analysis. Ki values are given in the table as means ± SEM.