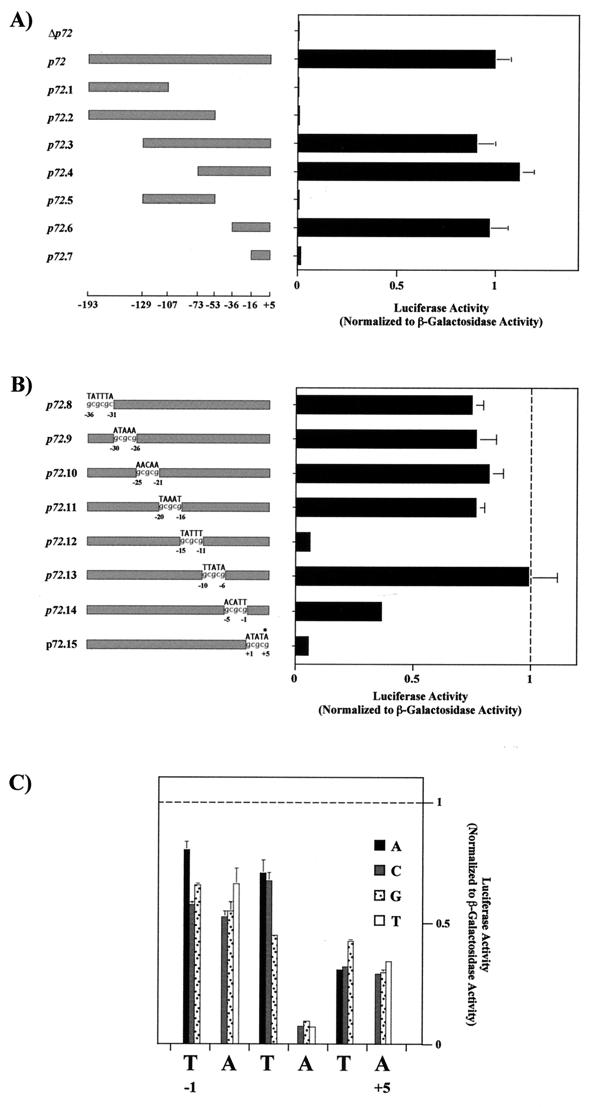
FIG. 2.
(A) Mapping of the p72 late promoter. Luciferase activity was measured in ASFV-infected Vero cells previously transfected with plasmids containing deletion fragments of the p72 promoter and was normalized to β-galactosidase activity produced from cotransfected control plasmid p72GAL10T. Transfections were carried out as described in the legend to Fig. 1 by using 12 ng of luciferase plasmids and 1 μg of p72GAL10T plasmid per 1.5 × 105 cells. Cells were infected with the BA71V strain of ASFV at a multiplicity of infection of 5 PFU per cell, and they were washed and harvested at 18 h postinfection. Luciferase and β-galactosidase activities were determined using the Dual-Light kit from Tropix Inc. The data are represented as the average plus the standard deviation of three assays and are normalized to p72 activity. The positions of the deletion fragment ends with respect to the transcriptional start point (+1) are indicated below the diagram. Δp72 corresponds to the luciferase gene without the p72 promoter. (B) Linker scanning analysis of the p72 promoter. The substituted sequence is indicated above each promoter, and the positions of mutated residues are indicated. Activities were determined as in panel A. All values were normalized to p72.6 activity, indicated with a vertical dashed line. The asterisk indicates the position of the first nucleotide of the p72 gene translation start codon. (C) Effect of single-nucleotide substitutions at or near the transcriptional start site. Activities were determined as for panel A. All values were normalized to p72.6 activity, indicated with a horizontal dashed line. Symbols for base substitutions are shown.