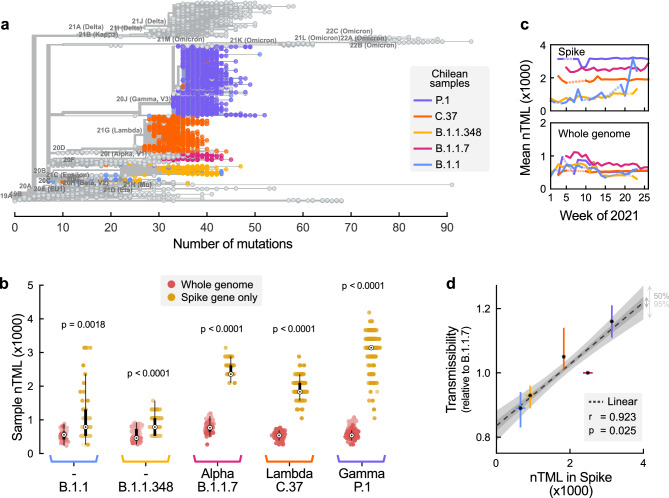
Figure 2.
Predominant variants are enriched with mutations in the Spike gene. (a) The Nextclade-based (https://clades.nextstrain.org/tree) phylogenetic tree of the SARS-CoV-2 variants isolated in Chile, visualised using Auspice online tool (https://auspice.us/ ) based on n = 2650 SARS-CoV-2 cases. The sequences are placed on a global reference tree (grey brunches and nodes), and clades are assigned to the nearest neighbour, while the branches with coloured circles represent lineages from Chile. (b) The normalised Total Mutational Load (nTML) indicates that the Spike gene is enriched in mutations compared to the entire genome for all analysed variants. The apparent discreteness of the Spike nTML traces is due to the shorter gene length. The white points denote the median, black boxes denote the interquartile ranges, and whiskers (thin black lines) extend until at most 1.5 times the length of the interquartile range, and dot opacity denotes the time when samples were collected (light old, dark recent). Significance levels were determined with an u-test, see Supplementary Table S2). c. The most predominant variants do not show a considerable drift in their average nTML over time. Dotted lines account for weeks when the variants were not observed. d. There is a marked and significant positive correlation between nTML in Spike and the variants’ relative transmissibility (median r = 0.923, p-value = 0.025). Vertical error bars are those reported in Figure 1, asterisks denote median values, and horizontal error bars were estimated through bootstrapping.