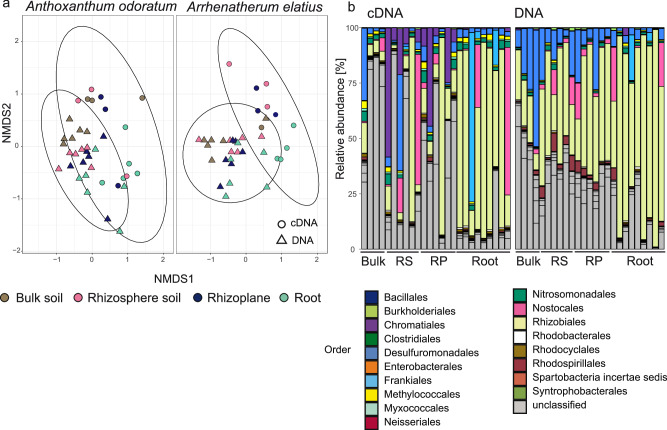
Fig. 4. Composition of active (cDNA-based) and DNA-based diazotroph communities associated with grasses.
a Non-metric multidimensional scaling (NMDS) ordination plots of nifH genes and transcripts (cDNA) in bulk soil, rhizosphere soil, rhizoplane, and roots of the grass species Anthoxanthum odoratum and Arrhenatherum elatius in the unfertilized treatment based on Bray–Curtis metric (stress value = 0.20). Ellipses indicate the 95% confidence interval. Exact sample sizes cDNA: bulk soil (n = 4 biologically independent samples), rhizosphere (n = 6 biologically independent samples), rhizoplane (n = 6 biologically independent samples), root (n = 9 biologically independent samples. Exact sample sizes DNA: for bulk soil, rhizosphere, rhizoplane, root (n = 12 biologically independent samples). b Taxonomic composition of the active diazotroph community (cDNA) and the DNA-based diazotroph community associated with bulk soil, rhizosphere, rhizoplane and roots of both grasses of the unfertilized treatment. Stacked bars reflect relative abundances (%) and are colored based on order level. Exact sample sizes cDNA and DNA: bulk soil (n = 4 biologically independent samples), rhizosphere (n = 6 biologically independent samples), rhizoplane (n = 6 biologically independent samples), root (n = 9 biologically independent samples). Bulk bulk soil, RS rhizosphere soil, RP rhizoplane.