ABSTRACT
Biofilm formation is an important virulence factor for methicillin-resistant Staphylococcus aureus (MRSA). The extracellular matrix of MRSA biofilms contains significant amounts of double-stranded DNA that hold the biofilm together. MRSA cells secrete micrococcal nuclease (Nuc1), which degrades double-stranded DNA. In this study, we used standard methodologies to investigate the role of Nuc1 in MRSA biofilm formation and dispersal. We quantified biofilm formation and extracellular DNA (eDNA) levels in broth and agar cultures. In some experiments, cultures were supplemented with sub-MIC amoxicillin to induce biofilm formation. Biofilm erosion was quantitated by culturing biofilms on rods and enumerating detached colony-forming units (CFUs), and biofilm sloughing was investigated by perfusing biofilms cultured in glass tubes with fresh broth and measuring the sizes of the detached cell aggregates. We found that an MRSA nuc1− mutant strain produced significantly more biofilm and more eDNA than a wild-type strain, both in the absence and presence of sub-MIC amoxicillin. nuc1− mutant biofilms grown on rods detached significantly less than wild-type biofilms. Detachment was restored by exogenous DNase or complementing the nuc1− mutant. In the sloughing assay, nuc1− mutant biofilms released cell aggregates that were significantly larger than those released by wild-type biofilms. Our results suggest that Nuc1 modulates biofilm formation, biofilm detachment, and the sizes of detached cell aggregates. These processes may play a role in the spread and subsequent survival of MRSA biofilms during biofilm-related infections.
IMPORTANCE
Infections caused by antibiotic-resistant bacteria known as methicillin-resistant Staphylococcus aureus (MRSA) are a significant problem in hospitals. MRSA forms adherent biofilms on implanted medical devices such as catheters and breathing tubes. Bacteria can detach from biofilms on these devices and spread to other parts of the body such as the blood or lungs, where they can cause life-threatening infections. In this article, researchers show that MRSA secretes an enzyme known as thermonuclease that causes bacteria to detach from the biofilm. This is important because understanding the mechanism by which MRSA detaches from biofilms could lead to the development of procedures to mitigate the problem.
KEYWORDS: Nuc1, Nuc2, sub-MIC, thermonuclease, clumping dispersal
INTRODUCTION
Staphylococcus aureus is a major public health burden in both community and hospital settings (1). In hospitals, S. aureus is a major cause of infections associated with implanted medical devices because of the ability of most S. aureus strains to form biofilms on biomaterial surfaces (2). S. aureus biofilms on implanted devices can act as a nidus of infection when bacterial cells detach from the biofilm and spread to other parts of the body such as the blood, lung, or bladder (3). In addition, nearly half of all S. aureus hospital isolates are methicillin-resistant S. aureus (MRSA), a multidrug-resistant strain that further complicates treatment of these common nosocomial infections (4).
S. aureus secretes numerous virulence factors into its surrounding environment, which contribute to host colonization and virulence. These include extracellular enzymes, pore-forming toxins, and superantigens (5). Among these is micrococcal nuclease (Nuc1), also known as thermonuclease, a secreted, Ca2+-dependent, thermostable phosphodiesterase that hydrolyzes both RNA and DNA (6). Nuc1 is produced by all strains of S. aureus. nuc1− mutant strains exhibit decreased survival in mouse models of peritonitis (6) and intranasal infection (7). Nuc1 may contribute to virulence in vivo by facilitating escape from neutrophil extracellular traps (7–9). Nuc1, along with other exoenzymes such as proteases, lipases, and hyaluronidases, may also contribute to tissue invasion or nutrient acquisition in vivo (10). All strains of S. aureus also produce a second extracellular nuclease termed Nuc2 (11, 12). Like Nuc1, the homologous Nuc2 enzyme is Ca2+-dependent, thermostable, and is able to hydrolyze both DNA and RNA (11, 12). Nuc2 is expressed in vivo, and purified Nuc2 protein inhibits S. aureus biofilm formation in microtiter plate wells and detaches preformed biofilms (11), suggesting that it may also play a role in biofilm formation and dispersal. However, nuc1 and nuc2 genes are located in different regions of the S. aureus chromosome, and the Nuc2 protein remains bound to the cell surface (11, 12).
Proteins and double-stranded DNA are the most common extracellular components of biofilms produced by most MRSA strains (13). Extracellular DNA (eDNA) consists of genomic DNA released by lysis of a subpopulation of cells within the biofilm (3, 14). eDNA may help stabilize the biofilm matrix by binding to secreted eDNA-binding proteins and membrane-attached lipoproteins (15) or to extracellular poly-N-acetylglucosamine polysaccharides (16). Previous studies have shown that biofilms produced by nuc1− mutant strains contain more eDNA than wild-type (WT) biofilms (17) and that biofilm formation is enhanced in nuc1− mutant strains (14, 17). In addition, expression of nuc1 is repressed during biofilm formation (6, 17). These findings suggest that exogenous Nuc1 can degrade eDNA, thereby decreasing the adhesive properties of the biofilm matrix.
In the present study, we investigated the role of Nuc1 and Nuc2 in biofilm formation using the MRSA strain LAC, a USA300 clone. By comparing a WT strain to nuc1−, nuc2−, and nuc1−/nuc2− mutant strains, we found that Nuc1, but not Nuc2, modulates the amount of eDNA in the biofilm matrix, the adhesiveness of the biofilms, and the amount of the biofilm formed, which is induced by low-dose amoxicillin. We further investigated the role of Nuc1 in two different mechanisms of biofilm dispersal, namely, erosion and sloughing. Biofilm erosion refers to the release of single cells or small clusters of cells from a biofilm, whereas sloughing refers to the detachment of large portions of the biofilm (18). Results from assays comparing biofilm dispersal in WT and nuc1− mutant strains suggests that Nuc1 not only mediates detachment of cells from MRSA biofilms but also modulates the sizes of the detached cell aggregates.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions
The bacterial strains used in this study are listed in Table 1. Bacteria were cultured on Tryptic Soy Agar plates or in filter-sterilized tryptic soy broth (BD Diagnostics). All cultures were incubated at 37°C. For plasmid-harboring strains, media were supplemented with 10 µg/mL chloramphenicol. To induce biofilm formation, media were supplemented with sub-MIC concentrations of amoxicillin ranging from 0.002 to 2 µg/mL. To induce biofilm detachment, the broth was supplemented with 10 µg/mL recombinant human DNase I (Genentech), which was previously shown to efficiently detach S. aureus biofilms from polystyrene microtiter plate wells (19). Bacterial inocula were prepared in sterile broth from 24-h-old agar colonies and then passed through a 5-µm-pore-size syringe filter to enrich for single cells, as previously described (20).
TABLE 1.
S. aureus strains used in this work
| Strain | Relevant characteristics | Reference |
|---|---|---|
| AH1263 | WT CA-MRSA USA300 strain LAC (ErmS variant) | (21) |
| AH1680 | AH1263 nuc1 (Nuc1− mutant) | (17) |
| AH3057 | AH1263 nuc2 (Nuc2− mutant) | (11) |
| AH3051 | AH1263 nuc1 nuc2 (Nuc1−/Nuc2− double mutant) | (11) |
| AH1787 | AH1680 carrying plasmid pCM28 (empty vector control) | (7) |
| AH1773 | AH1680 carrying plasmid pCM28nuc1 (Nuc1 complementing vector) | (7) |
Six-well microtiter plate biofilm assay
Aliquots of inocula (4 mL each; ca. 102 CFU/mL) were transferred to the wells of a 6-well microtiter plate (Falcon #353046), and the plate was incubated statically under conditions of low vibration, as previously described (22). After 18 h, a 1 cm2 area in the center of each well was photographed. Wells were then rinsed with water and stained for 1 min with 4 mL of Gram’s crystal violet. Wells were rinsed with water and dried, and the same area of each well was rephotographed.
Ninety-six-well microtiter plate biofilm assay
Aliquots of inocula (180 µL each; 105–106 CFU/mL) were transferred to the wells of a 96-well microtiter plate (Corning #3599) containing 20 µL of an antibiotic dissolved in water at a concentration equal to 10 times the desired final concentration. Control wells were filled with 180 µL of the inoculum and 20 µL of water or 180 µL of sterile broth and 20 µL of water. The plates were incubated for 18 h. The amount of biofilm biomass in each well was quantitated by rinsing the wells with water and staining for 1 min with 200 µL of Gram’s crystal violet. The wells were then rinsed with water and dried. The bound crystal violet was dissolved in 200 µL of 33% acetic acid and quantitated by measuring the absorbance of the wells at 620 nm.
Isolation and analysis of extracellular DNA
Extracellular DNA was isolated from lawns of bacteria cultured on agar, also known as colony biofilms, using the method described by Karwacki et al. (23). Briefly, 100-µL aliquots of the inoculum (>108 CFU) was spread onto agar plates supplemented with 0 or 0.2 µg/mL amoxicillin. After incubation for 24 h, the cell paste from each plate was transferred to a separate, preweighed 1.5-mL microcentrifuge tube, weighed, and resuspended in TE buffer at a concentration of 1 mg/mL. The sample was mixed by vortex agitation for 10 min, and the cells were pelleted by centrifugation. The supernatant was sterilized by passage through a 0.2-μm-pore-size filter. A 20 µL volume of the colony biofilm extract was analyzed by agarose gel electrophoresis, and the DNA was visualized by staining with ethidium bromide.
Biofilm erosion assay
An apparatus to measure biofilm erosion under static conditions (Fig. 1A) was constructed and employed as follows. First, a 2-mm-diameter hole was drilled in the center of a 35-mm-diam Petri dish lid (Sarstedt # 83.1800.001). Next, a 100 mm polystyrene or glass rod was placed in the hole and secured in a vertical position with hot-melt adhesive. The polystyrene rods (1.5 mm diameter) were purchased from Plastruct (City of Industry CA), and the glass rods were made from 1.9-mm-diameter open-one-end capillary tubes (Stuart Glass # EW-03013-64) with the closed end facing down. The rods were positioned so that 5 mm protruded from the top of the lid (Fig. 1A). The apparatus were sterilized with 70% ethanol, air-dried, and then placed on top of 50-mL conical centrifuge tubes (Corning # 430290) containing 20 mL of the bacterial inoculum at 105–106 CFU/mL. Approximately 30 mm of the rod or capillary tube was suspended in the broth. After incubation for 16 h, the apparatus were carefully transferred to fresh tubes containing 25 mL of prewarmed sterile broth and incubated for 1 min to detach loosely adherent cells. The apparatus were then transferred to fresh tubes containing 25 mL of prewarmed sterile broth. After 2 h, the apparatus were removed from the centrifuge tubes. To enumerate the CFUs in the broth, the broth was mixed by vortex agitation for 30 s, serially diluted in phosphate-buffered saline (PBS), and then plated on agar for CFU enumeration. To enumerate the CFUs on the rods, the rods were detached from the Petri dish lids, placed in 15-mL conical centrifuge tubes containing 10 mL of PBS, and sonicated for 30 s at 40% duty cycle in an ultrasonic homogenizer equipped with a 1/8 in. probe (Fisher Scientific model 505). Sonicates were serially diluted in PBS and plated on agar for CFU enumeration. The fraction of detached cells (percent CFUs in broth) was calculated by dividing the total number of CFUs in the broth by the total number of CFUs in the broth plus the total number of CFUs on the rod. Assays were performed in duplicate to quadruplicate tubes for each experimental condition.
Fig 1.
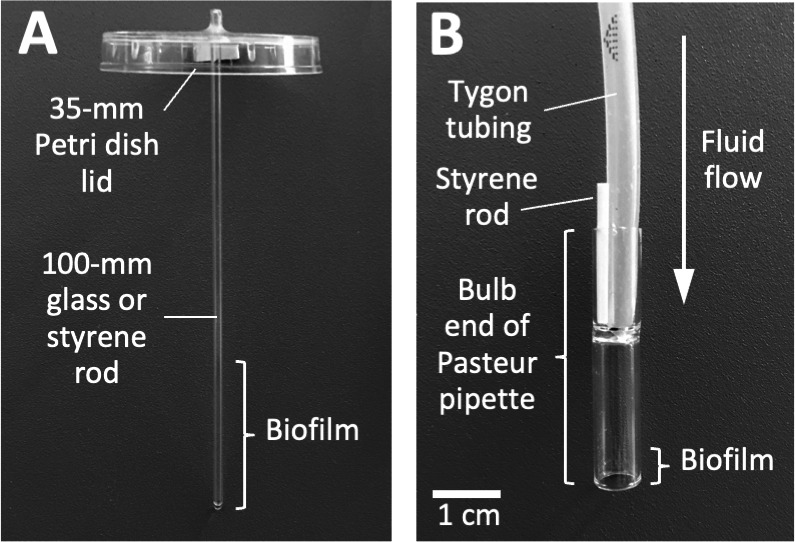
Apparatus used to measure biofilm erosion (A) and biofilm sloughing (B). The construction and use of these apparatus are presented in Materials and Methods.
Biofilm sloughing assay
An apparatus to measure biofilm sloughing under flow conditions (Fig. 1B) was constructed and employed as follows. First, biofilm reactors were constructed by cutting 38-mm-long sections from the bulb ends of standard 9-in. flint glass Pasteur pipettes (VWR # 14672-380) by using a glass cutter. The glass reactors were rinsed in 1% HCl for 2 h, rinsed with 70% ethanol and ultrapure water, and then air-dried. Glass reactors were inoculated by standing them (bulb-side-down) in a 6-well microtiter plate well containing 2 mL of the bacterial inoculum at 105–106 CFU/mL, which brought the bottom 5 mm of each reactor in contact with the inoculum. After 10 min, the reactors were connected to 1/8 × 3/16 in. silicon tubing (Tygon 3350 #ABW00006) and perfused with fresh broth for 48 h at a rate of 5–7 mL/h. A short 1.5-mm-diameter polystyrene rod (Fig. 1B) functioned to secure the tubing to the glass reactor and to form a vent that allowed the reactor to act as a continuous drip chamber. A total of four reactors were perfused in each experiment, two inoculated with the WT strain and two with the nuc1− mutant strain. To enumerate CFUs in the eluate, drops were collected directly into 1.5-mL microcentrifuge tubes, mixed by vortex agitation for 30 s, serially diluted in PBS, and plated on agar for CFU enumeration. To measure the sizes of the detached cell aggregates, drops were collected directly onto glass microscope slides and then fixed, stained with Gram’s crystal violet, and photographed at 400× under an inverted microscope. For each strain, the distribution of particle sizes in five random microscopic fields was analyzed using ImageJ software. For direct visualization of detached cell aggregates, drops of the eluate were collected into the wells of a 6-well microtiter plate and then photographed on a dark background.
Statistics and reproducibility of results
Biofilm erosion assays were performed in triplicate tubes or quadruplicate tubes and were repeated two to four times. The significance of differences between percent CFUs in broth values was calculated using a two-tailed Student’s t-test for pairwise comparisons and a one-way ANOVA with Tukey’s post hoc analysis for comparison of more than two groups. A P value of <0.05 was considered significant. Biofilm sloughing assays were performed in duplicate reactors for each strain (wild-type and nuc1−) and were repeated twice. The significance of differences between particle size distributions in the sloughing assay was calculated using a two-way chi squared test.
RESULTS
Biofilm formation by WT and nuclease mutant strains in 6-well microtiter plate wells
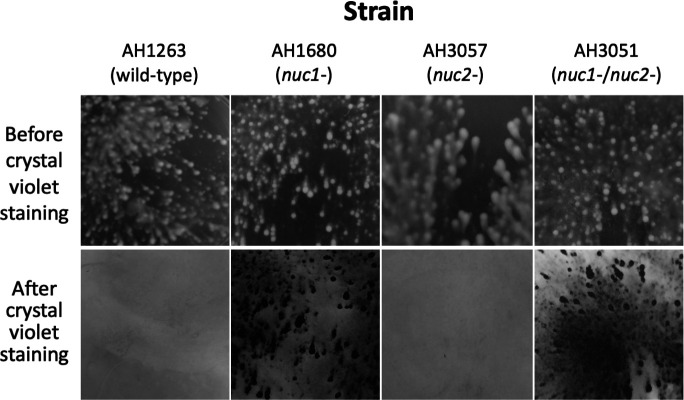
MRSA WT, nuc1−, nuc2−, and nuc1−/nuc2− strains were cultured in broth in 6-well microtiter plates at low densities and under conditions of low vibration. After 18 h, the wells were photographed (Fig. 2, top panels). All four strains produced surface-attached biofilms under these conditions. nuc1− and nuc1−/nuc2− biofilms appeared denser and more compact than those of the WT and nuc2− strains. After rinsing the wells with water and staining with crystal violet, biofilms of the nuc1− and nuc1−/nuc2− strains remained firmly attached to the surface, whereas biofilms of the WT and nuc2− strains were readily detached (Fig. 2, bottom panels). These results suggest that Nuc1, but not Nuc2, contributes to MRSA USA300 biofilm formation under these conditions.
Fig 2.
Biofilm formation by MRSA USA300 WT and nuclease mutant strains in 6-well tissue culture-treated microplates under conditions of low vibration. Top panels show surface-attached growth after incubation for 16 h. Bottom panels show the identical areas of the plate after they were rinsed with water and stained with crystal violet. The size of each imaged area is 1 cm2. The 6-well microplate assay was performed three times using four to six wells for each strain in each experiment. All four strains exhibited the same biofilm growth and crystal violet detachment phenotypes in all wells in each experiment. Representative wells are shown above.
Induction of biofilm formation by sub-MIC amoxicillin in WT and nuclease mutant strains
Previous studies showed that low doses of amoxicillin induce biofilm formation in MRSA USA300 by a mechanism that involved autolysis and eDNA release (24, 25). To determine whether Nuc1 or Nuc2 plays a role in this process, we used a 96-well crystal violet binding assay to quantitate biofilm formation by WT, nuc1−, nuc2−, and nuc1−/nuc2− strains cultured in sub-MIC amoxicillin concentrations ranging from 0.002 to 2 µg/mL (Fig. 3). The MIC of amoxicillin against strain USA300 is >8 µg/mL (24). Low-dose amoxicillin induced biofilm formation in all four strains, with maximum induction occurring at a concentration of 0.2 µg/mL. However, biofilm induction occurred over a greater range of amoxicillin concentrations and resulted in a significantly greater amount of biofilm biomass in the nuc1− and nuc1−/nuc2− strains compared to the WT and nuc2− strains (Fig. 3). These results suggest that Nuc1, but not Nuc2, can modulate the structure of MRSA biofilms under both antibiotic-induced and uninduced conditions. These results are consistent with those of previous studies showing that nuclease activity is not required for β-lactam-induced MRSA biofilm formation (24).
Fig 3.
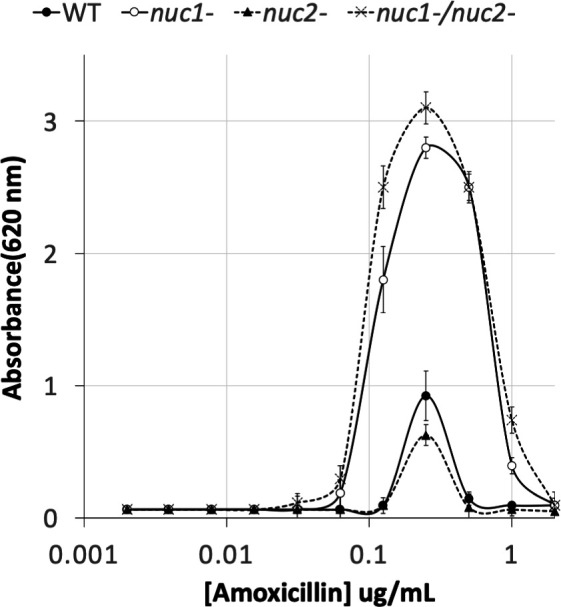
Biofilm formation by MRSA USA300 WT and nuclease mutant strains in 96-well microtiter plates in the presence of increasing concentrations of sub-MIC amoxicillin. Absorbance at 620 nm is proportional to biofilm biomass. Values show mean and range for duplicate wells. The biofilm induction assay was performed three times using duplicate wells for each strain. Duplicate wells exhibited an average variation of 7% within each experiment and an average of 18% between experiments. In all three experiments, the dose–response curves (antibiotic concentration, amplitude of stimulus response, and range of the stimulatory dose) were the same for all four strains tested. Representative graphs from one experiment are shown above.
eDNA production in WT and nuclease mutant strains
To investigate the role of Nuc1 and Nuc2 in MRSA USA300 eDNA production, we isolated eDNA from colony biofilms of WT, nuc1−, nuc2−, and nuc1−/nuc2− strains and isolated the DNA by agarose gel electrophoresis (Fig. 4A). The nuc1− and nuc1−/nuc2− strains produced significantly more high-molecular weight eDNA (> 25 kb) than the WT and nuc2− strains. All four strains produced high-molecular weight DNA in the presence of low-dose amoxicillin (Fig. 4B), which suggests that Nuc2 may regulate eDNA production under antibiotic-induced conditions. Taken together, our results demonstrate that high-molecular weight eDNA production inversely correlates with Nuc1 production (Fig. 4), but positively correlates with biofilm tenacity (Fig. 2) and antibiotic-induced biofilm formation (Fig. 3).
Fig 4.
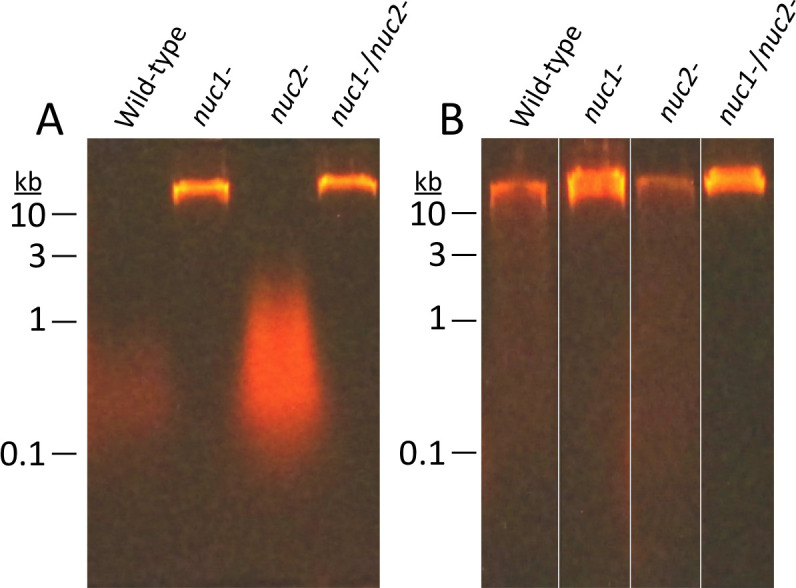
Agarose gel electrophoretic analysis of extracellular DNA (eDNA) produced by MRSA USA300 WT and nuclease mutant strains. eDNA was harvested from colony biofilms cultured in TSA (A) or TSA supplemented with 0.2 µg/mL amoxicillin (B). Numbers at the left indicate the sizes of molecular weight markers electrophoresed in an adjacent lane. eDNA was isolated and analyzed from three independent cultures of each strain for each condition (±antibiotic). In all cases, the amount and size of eDNA and its relative abundance among the strains exhibited the same pattern as those in the representative gel images shown above.
Role of Nuc1 in biofilm erosion
We next investigated the role of Nuc1 in MRSA biofilm erosion. We focused on Nuc1 because it is the primary modulator of MRSA eDNA production and biofilm formation under uninduced conditions. Biofilm erosion was quantitated by culturing WT and nuc1− biofilms on polystyrene or glass rods. After 2 h, the rods were removed from the tubes and the number of CFUs on the rod and in the broth was measured. Percent biofilm dispersal was calculated as (broth CFUs)/(broth CFUs + rod CFUs) ×100. The total CFUs per tube ranged from 108 to 109 for both polystyrene and glass rods.
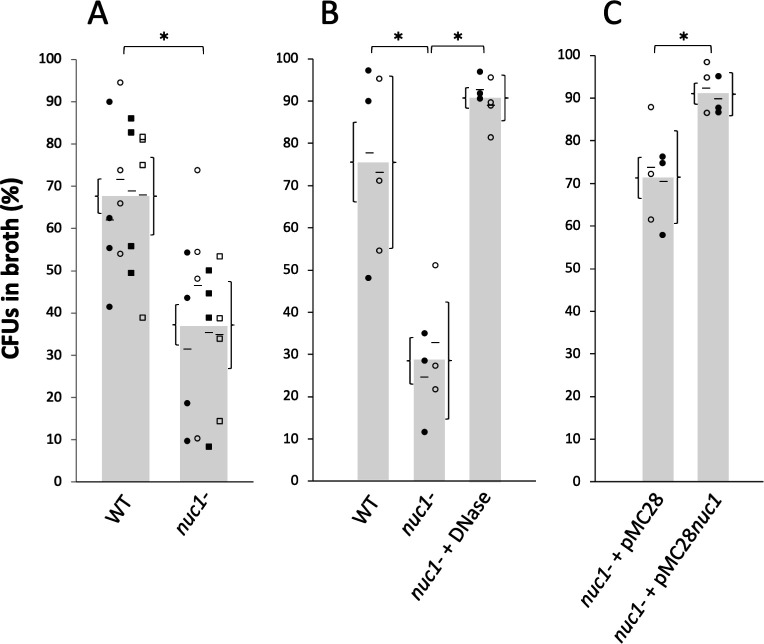
Figure 5A shows the results from multiple biofilm erosion assays performed on polystyrene rods. On average, significantly more CFUs detached from WT biofilms compared to the number of CFUs that detached from nuc1− mutant biofilms (68% of CFUs in the broth for WT versus 38% for nuc1− mutant). The same erosion phenotype was observed when biofilms were cultured on glass rods (76% CFUs in broth for the WT versus 38% for nuc1− strains) (Fig. 5B). There was no significant difference between erosion rates from polystyrene or glass rods for either the WT strain (P = 0.37) or the nuc1− mutant strain (P = 0.37). However, significantly more number of CFUs were present in the broth when nuc1− mutant biofilms were allowed to erode from glass rods in broth supplemented with DNase compared to the no DNase control (Fig. 5B) or when the nuc1− strain carried a WT nuc1 gene on the plasmid. (Fig. 5C). These findings confirm that Nuc1 mediates MRSA biofilm erosion.
Fig 5.
Erosion of MRSA USA300 wild-type (WT) and nuc1− mutant biofilms from polystyrene rods (A) and glass rods (B and C). Bars show the average percent of total CFUs in the broth after 2 h. Each symbol represents one rod. Results from individual experiments (four in panel A and two in panels B and C) are indicated by different symbols (filled and open circles and squares). Short horizontal bars indicate mean values for individual experiments. For panel A, there was a significant difference between the means of the four independent experiments, as determined by t-test (P = 0.0002), and no significant difference between the means of any pair of individual experiments for either the WT or nuc1− strains, as determined by one-way ANOVA with Tukey’s post hoc analysis (P = 0.889 for the WT strain and P = 0.743 for the nuc1− strain). Curly brackets to the left and right of the value bars indicate SE and 95% CI in panel A, respectively, and SE and SD in panels B and C, respectively. Asterisks denote groups with significantly different distributions (P < 0.001) based on t-tests for panels A and C and an ANOVA test for panel B. (A) Erosion of WT and nuc1− biofilms from polystyrene rods. (B) Erosion of WT and nuc1− biofilms from glass rods. Some nuc1− biofilms were incubated in broth supplemented with DNase. (C) Erosion of nuc1− biofilms harboring pMB28 (empty plasmid control) or pMB28nuc1 (nuc1 complementing plasmid) from glass rods.
Role of Nuc1 in biofilm sloughing
Biofilm sloughing is the detachment of large portions of a biofilm, which usually occurs in the later stages of biofilm formation (18). To quantitate biofilm sloughing, we grew WT and nuc1− mutant biofilms in glass tubes perfused with the fresh broth and measured the number of CFUs and sizes of detached cell aggregates in the eluate (Fig. 1B). Both strains formed biofilms in the glass tube reactors and dispersed. Over the course of the 48-h assay, the nuc1− mutant biofilm grew inside the silicone tubing that supplied fresh broth to the reactor as evidenced by crystal violet staining (Fig. 6A). Biofilms of both strains dispersed at high levels from 18 to 52 h (Fig. 6B), but nuc1− mutant biofilms released at least ten times more CFUs than WT biofilms at all time points tested, probably because of a larger amount of biofilm biomass.
Fig 6.
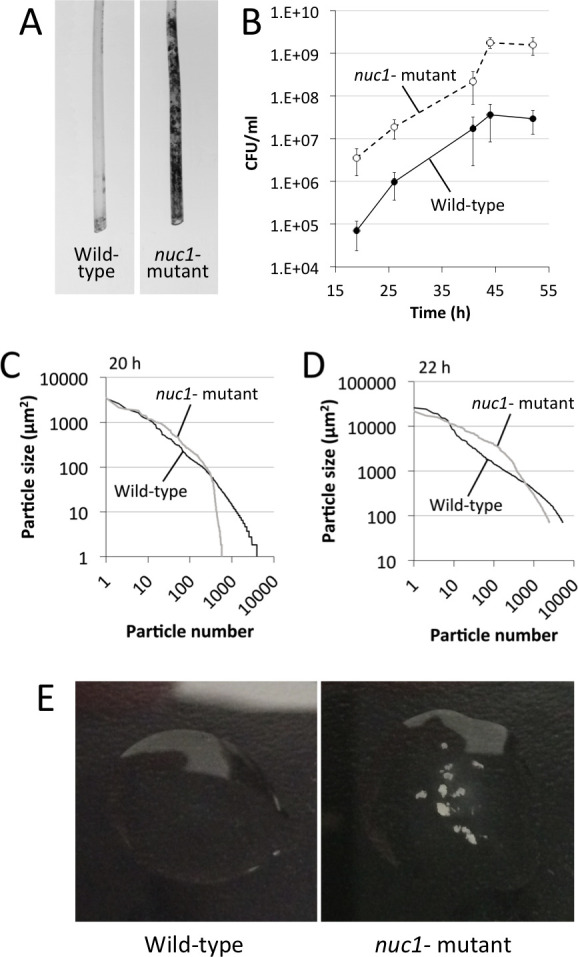
Biofilm sloughing assay. (A) Photographs of the sections of silicone tubing located directly above the glass biofilm reactors. The tubing sections were stained with crystal violet after 48 h of perfusion. (B) CFU/mL in the flowthrough from 17 to 52 h. Values show the mean and range for duplicate reactors. (C and D) Distribution of particle sizes in the eluates from two independent sloughing experiments. Eluates were analyzed after 20 h (C) or 22 h (D) of perfusion. (E) Photographs of drops of the eluate after 48 h of perfusion.
We also measured the sizes of particles that detached from WT and nuc1− mutant biofilms using ImageJ software. Graphs showing the distribution of particle sizes from two independent sloughing experiments are shown in Fig. 6C and D. Characteristics of the detached particles from both experiments are summarized in Table 2. In both experiments, the distributions of particle sizes were significantly different (P < 0.0001; two-way chi squared test) (Fig. 6C and D). In addition, the average number of particles in the nuc1− eluate was significantly less than the number in the WT eluate, and the average particle size in the nuc1− eluate was significantly greater than that in the WT eluate in both experiments (Table 2). Differences between the number of particles, the average particle size, and the range of particle sizes between the two experiments shown in Fig. 6C and D and Table 2 may be due to the 2-h time difference or due to variations in the way in which the cells were fixed and stained or the manner in which the images were captured and analyzed on different days. After 48–52 h of perfusion, macroscopic clusters of cells were present in the nuc1− eluate, but not in the WT eluate (Fig. 6E).
TABLE 2.
Characteristics of cell aggregates in the eluate of WT and nuc1− biofilms perfused with the broth for 20–22 ha
| Expt 1 (20 h) | Expt 2 (22 h) | |||
|---|---|---|---|---|
| AH1263 (WT) |
AH1680 (nuc1−) |
AH1263 (WT) |
AH1680 (nuc1−) |
|
| Average number of particles (± SD) per microscopic field | 1,436 ± 438 |
135 ± 33b |
1,072 ± 287 |
492 ± 108b |
| Average particle size (µm2) | 15 | 144b | 317 | 723b |
Results from two independent experiments are shown.
Significantly different from strain AH1263 (P < 0.01, two-tailed Student’s t-test).
DISCUSSION
Previous studies showed that biofilms produced by MRSA nuc1− mutant strains are thicker and contain more eDNA than WT biofilms (14, 17, 25) and that nuc1 expression is repressed during biofilm formation (6, 17). These findings are consistent with the hypothesis that Nuc1 degrades eDNA and decreases the adhesive properties of the biofilm matrix. A corollary hypothesis is that Nuc1 may function as an endogenous mediator of biofilm dispersal in this species. Dispersal of biofilms from indwelling medical devices is a clinically relevant process that may lead to invasive infections such as ventilator-associated pneumonia and endocarditis (26). Although Moormeier et al. (27) showed that Nuc1 mediates the detachment and dispersal of cells prior to the development of characteristic tower-like structures in the early stages of S. aureus biofilm formation, no studies have directly tested the hypothesis that Nuc1 mediates the detachment and dispersal of cells from mature S. aureus biofilms.
In the present study, we constructed apparatus to measure S. aureus biofilm erosion and biofilm sloughing. Erosion refers to the continuous release of single cells or small clusters of cells from a biofilm at low levels over the course of biofilm formation, whereas sloughing refers to the sudden detachment of large portions of the biofilm, usually during the later stages of biofilm formation (18). We focused on Nuc1 because Nuc2 appears to play a minor role in MRSA biofilm formation (Fig. 3) and eDNA release (Fig. 4) under the conditions tested. Detachment by the nuc1− mutant was restored to WT levels by exogenous DNase (Fig. 5B) or a WT nuc1 gene on a plasmid (Fig. 5C), confirming that Nuc1 mediates biofilm erosion under the conditions tested. In the biofilm sloughing assay, the nuc1− mutant released ten times as many CFUs as the WT strain (Fig. 6B), likely due to the increased biofilm biomass in these strains. The nuc1− mutant strain also released significantly larger cell aggregates than the WT strain (Fig. 6C and D; Table 2). The sizes of the detached cell aggregates in our study were similar to the sizes of aggregates that detached from S. aureus biofilms cultured in glass tubes perfused with the brain heart infusion medium (28) and in silicone tubing perfused with human plasma (29). Large, detached aggregates of S. aureus cells were previously shown to be highly resistant to antibiotics (28, 30) and phagocytosis (31, 32) and capable of initiating the colonization of endothelial cell layers under flow (29), which suggests that biofilm sloughing may contribute to metastatic infections associated with S. aureus (28).
Our results demonstrate that Nuc1 modulates biofilm erosion and sloughing in S. aureus USA300. A homologous nuclease was shown to modulate eDNA production, biofilm dispersal, and biofilm aggregate size in nontypeable Haemophilus influenzae (33). Since eDNA is a structural component of Pseudomonas aeruginosa biofilms (34), it is possible that Nuc1 may passively contribute to P. aeruginosa biofilm formation and dispersal during S. aureus/P. aeruginosa coinfection in the airways of cystic fibrosis patients (35) and in chronic wounds (36).
ACKNOWLEDGMENTS
The authors thank Aleksandr Florjanczyk, Maria Ochiai, Caleb Jones, Gabrielle Kyle-Lion, Roxanna Stapleton, and Amelia Crabtree (American University) for technical assistance.
This work was supported in part by NIH NIAID grant AI083211 to A.R.H.
Contributor Information
Alexander R. Horswill, Email: alexander.horswill@cuanschutz.edu.
Sarah E. F. D'Orazio, University of Kentucky College of Medicine, Lexington, Kentucky, USA
REFERENCES
- 1. Percival SL, Suleman L, Vuotto C, Donelli G. 2015. Healthcare-associated infections, medical devices and biofilms: risk, tolerance and control. J Med Microbiol 64:323–334. doi: 10.1099/jmm.0.000032 [DOI] [PubMed] [Google Scholar]
- 2. Schilcher K, Horswill AR. 2020. Staphylococcal biofilm development: structure, regulation, and treatment strategies. Microbiol Mol Biol Rev 84:e00026-19. doi: 10.1128/MMBR.00026-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Archer NK, Mazaitis MJ, Costerton JW, Leid JG, Powers ME, Shirtliff ME. 2011. Staphylococcus aureus biofilms: properties, regulation and roles in human disease. Virulence 2:445–459. doi: 10.4161/viru.2.5.17724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Diekema DJ, Pfaller MA, Shortridge D, Zervos M, Jones RN. 2019. Twenty-year trends in antimicrobial susceptibilities among Staphylococcus aureus from the SENTRY antimicrobial surveillance program. Open Forum Infect Dis 6:S47–S53. doi: 10.1093/ofid/ofy270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Busche T, Hillion M, Van Loi V, Berg D, Walther B, Semmler T, Strommenger B, Witte W, Cuny C, Mellmann A, Holmes MA, Kalinowski J, Adrian L, Bernhardt J, Antelmann H. 2018. Comparative secretome analyses of human and zoonotic Staphylococcus aureus isolates CC8, CC22, and CC398. Mol Cell Proteom 17:2412–2433. doi: 10.1074/mcp.RA118.001036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Olson ME, Nygaard TK, Ackermann L, Watkins RL, Zurek OW, Pallister KB, Griffith S, Kiedrowski MR, Flack CE, Kavanaugh JS, Kreiswirth BN, Horswill AR, Voyich JM. 2013. Staphylococcus aureus nuclease is an SaeRS-dependent virulence factor. Infect Immun 81:1316–1324. doi: 10.1128/IAI.01242-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Berends ETM, Horswill AR, Haste NM, Monestier M, Nizet V, von Köckritz-Blickwede M. 2010. Nuclease expression by Staphylococcus aureus facilitates escape from neutrophil extracellular traps. J Innate Immun 2:576–586. doi: 10.1159/000319909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bhattacharya M, Berends ETM, Zheng X, Hill PJ, Chan R, Torres VJ, Wozniak DJ. 2020. Leukocidins and the nuclease Nuc prevent neutrophil-mediated killing of Staphylococcus aureus biofilms. Infect Immun 88:e00372-20. doi: 10.1128/IAI.00372-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thammavongsa V, Missiakas DM, Schneewind O. 2013. Staphylococcus aureus degrades neutrophil extracellular traps to promote immune cell death. Science 342:863–866. doi: 10.1126/science.1242255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tam K, Torres VJ. 2019. Staphylococcus aureus secreted toxins and extracellular enzymes. Microbiol Spectr 7. doi: 10.1128/microbiolspec.GPP3-0039-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kiedrowski MR, Crosby HA, Hernandez FJ, Malone CL, McNamara JO, Horswill AR. 2014. Staphylococcus aureus Nuc2 is a functional, surface-attached extracellular nuclease. PLoS ONE 9:e95574. doi: 10.1371/journal.pone.0095574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tang J, Zhou R, Shi X, Kang M, Wang H, Chen H. 2008. Two thermostable nucleases coexisted in Staphylococcus aureus: evidence from mutagenesis and in vitro expression. FEMS Microbiol Lett 284:176–183. doi: 10.1111/j.1574-6968.2008.01194.x [DOI] [PubMed] [Google Scholar]
- 13. Sugimoto S, Sato F, Miyakawa R, Chiba A, Onodera S, Hori S, Mizunoe Y. 2018. Broad impact of extracellular DNA on biofilm formation by clinically isolated methicillin-resistant and -sensitive strains of Staphylococcus aureus. Sci Rep 8:2254. doi: 10.1038/s41598-018-20485-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mann EE, Rice KC, Boles BR, Endres JL, Ranjit D, Chandramohan L, Tsang LH, Smeltzer MS, Horswill AR, Bayles KW. 2009. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS ONE 4:e5822. doi: 10.1371/journal.pone.0005822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kavanaugh JS, Flack CE, Lister J, Ricker EB, Ibberson CB, Jenul C, Moormeier DE, Delmain EA, Bayles KW, Horswill AR. 2019. Identification of extracellular DNA-binding proteins in the biofilm matrix. mBio 10:e01137-19. doi: 10.1128/mBio.01137-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mlynek KD, Bulock LL, Stone CJ, Curran LJ, Sadykov MR, Bayles KW, Brinsmade SR. 2020. Genetic and biochemical analysis of CodY-mediated cell aggregation in Staphylococcus aureus reveals an interaction between extracellular DNA and polysaccharide in the extracellular matrix. J Bacteriol 202:e00593-19. doi: 10.1128/JB.00593-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kiedrowski MR, Kavanaugh JS, Malone CL, Mootz JM, Voyich JM, Smeltzer MS, Bayles KW, Horswill AR. 2011. Nuclease modulates biofilm formation in community-associated methicillin-resistant Staphylococcus aureus. PLoS ONE 6:e26714. doi: 10.1371/journal.pone.0026714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kaplan JB. 2010. Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J Dent Res 89:205–218. doi: 10.1177/0022034509359403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kaplan JB, LoVetri K, Cardona ST, Madhyastha S, Sadovskaya I, Jabbouri S, Izano EA. 2012. Recombinant human DNase I decreases biofilm and increases antimicrobial susceptibility in staphylococci. J Antibiot (Tokyo) 65:73–77. doi: 10.1038/ja.2011.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Izano EA, Amarante MA, Kher WB, Kaplan JB. 2008. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus Epidermidis biofilms. Appl Environ Microbiol 74:470–476. doi: 10.1128/AEM.02073-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boles BR, Thoendel M, Roth AJ, Horswill AR. 2010. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS ONE 5:e10146. doi: 10.1371/journal.pone.0010146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kaplan JB, Fine DH. 2002. Biofilm dispersal of Neisseria subflava and other phylogenetically diverse oral bacteria. Appl Environ Microbiol 68:4943–4950. doi: 10.1128/AEM.68.10.4943-4950.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Karwacki MT, Kadouri DE, Bendaoud M, Izano EA, Sampathkumar V, Inzana TJ, Kaplan JB. 2013. Antibiofilm activity of Actinobacillus pleuropneumoniae serotype 5 capsular polysaccharide. PLoS ONE 8:e63844. doi: 10.1371/journal.pone.0063844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kaplan JB, Izano EA, Gopal P, Karwacki MT, Kim S, Bose JL, Bayles KW, Horswill AR. 2012. Low levels of β-lactam antibiotics induce extracellular DNA release and biofilm formation in Staphylococcus aureus. mBio 3:e00198-12. doi: 10.1128/mBio.00198-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mlynek KD, Callahan MT, Shimkevitch AV, Farmer JT, Endres JL, Marchand M, Bayles KW, Horswill AR, Kaplan JB. 2016. Effects of low-dose amoxicillin on Staphylococcus aureus USA300 biofilms. Antimicrob Agents Chemother 60:2639–2651. doi: 10.1128/AAC.02070-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Adair CG, Gorman SP, Feron BM, Byers LM, Jones DS, Goldsmith CE, Moore JE, Kerr JR, Curran MD, Hogg G, Webb CH, McCarthy GJ, Milligan KR. 1999. Implications of endotracheal tube biofilm for ventilator-associated pneumonia. Intensive Care Med 25:1072–1076. doi: 10.1007/s001340051014 [DOI] [PubMed] [Google Scholar]
- 27. Moormeier DE, Bose JL, Horswill AR, Bayles KW. 2014. Temporal and stochastic control of Staphylococcus aureus biofilm development. mBio 5:e01341-14. doi: 10.1128/mBio.01341-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fux CA, Wilson S, Stoodley P. 2004. Detachment characteristics and oxacillin resistance of Staphyloccocus aureus biofilm emboli in an in vitro catheter infection model. J Bacteriol 186:4486–4491. doi: 10.1128/JB.186.14.4486-4491.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Grønnemose RB, Saederup KL, Kolmos HJ, Hansen SWK, Asferg CA, Rasmussen KJ, Palarasah Y, Andersen TE. 2017. A novel in vitro model for haematogenous spreading of S. aureus device biofilms demonstrating clumping dispersal as an advantageous dissemination mechanism. Cell Microbiol 19:12. doi: 10.1111/cmi.12785 [DOI] [PubMed] [Google Scholar]
- 30. Haaber J, Cohn MT, Frees D, Andersen TJ, Ingmer H. 2012. Planktonic aggregates of Staphylococcus aureus protect against common antibiotics. PLoS One 7:e41075. doi: 10.1371/journal.pone.0041075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alhede M, Lorenz M, Fritz BG, Jensen PØ, Ring HC, Bay L, Bjarnsholt T. 2020. Bacterial aggregate size determines phagocytosis efficiency of polymorphonuclear leukocytes. Med Microbiol Immunol 209:669–680. doi: 10.1007/s00430-020-00691-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pettygrove BA, Kratofil RM, Alhede M, Jensen PØ, Newton M, Qvortrup K, Pallister KB, Bjarnsholt T, Kubes P, Voyich JM, Stewart PS. 2021. Delayed neutrophil recruitment allows nascent Staphylococcus aureus biofilm formation and immune evasion. Biomaterials 275:120775. doi: 10.1016/j.biomaterials.2021.120775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cho C, Chande A, Gakhar L, Bakaletz LO, Jurcisek JA, Ketterer M, Shao J, Gotoh K, Foster E, Hunt J, O’Brien E, Apicella MA. 2015. Role of the nuclease of nontypeable Haemophilus influenzae in dispersal of organisms from biofilms. Infect Immun 83:950–957. doi: 10.1128/IAI.02601-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S, Molin S, Givskov M, Tolker-Nielsen T. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol 59:1114–1128. doi: 10.1111/j.1365-2958.2005.05008.x [DOI] [PubMed] [Google Scholar]
- 35. Wieneke MK, Dach F, Neumann C, Görlich D, Kaese L, Thißen T, Dübbers A, Kessler C, Große-Onnebrink J, Küster P, Schültingkemper H, Schwartbeck B, Roth J, Nofer J-R, Treffon J, Posdorfer J, Boecken JM, Strake M, Abdo M, Westhues S, Kahl BC. 2021. Association of diverse Staphylococcus aureus populations with Pseudomonas aeruginosa coinfection and inflammation in cystic fibrosis airway infection. mSphere 6:e0035821. doi: 10.1128/mSphere.00358-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ibberson CB, Whiteley M. 2020. The social life of microbes in chronic infection. Curr Opin Microbiol 53:44–50. doi: 10.1016/j.mib.2020.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]