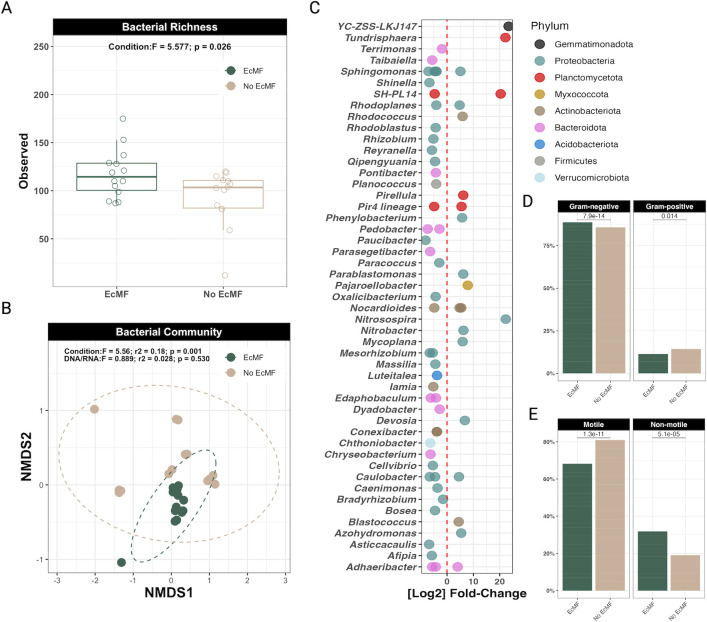
Fig 1.
Diversity estimates of bacterial communities either with EcMF or without EcMF. (A) Observed bacterial richness between treatments (i.e., either with EcMF or without EcMF [No EcMF]). (B) Beta diversity comparison between conditions and library preparation type (i.e., from DNA templates or from RNA to cDNA). Stress = 0.0914. In both panels A and B, n = 13 for each condition. (C) DESeq2 analysis showing which ASVs are differentially abundant in either the presence of EcMF (left of the red dotted line) or the absence of EcMF (right of the red dotted line). Each ASV is denoted by a single dot. ASVs are grouped by genus on the y-axis and colored by phylum designation. All the ASVs shown are statistically enriched (P < 0.001), and all P-values were corrected using the Benjamini and Hochberg method (52). (D) Ratios of Gram-negative and Gram-positive bacteria that were differentially abundant according to DESeq2. (E) Ratios of motile versus non-motile bacteria that were differentially abundant according to DESeq2. P values are shown above each comparison (t-test, where P < 0.05 equals a significant difference between groups).