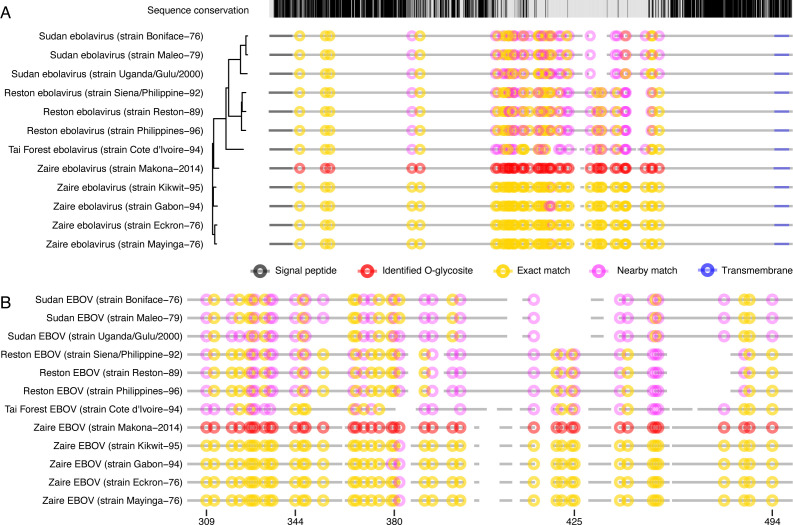
Fig 4.
Conservation of O-glycosylated amino acids. Clustal Omega server was used to align amino acid sequences of reviewed UniProtKB entries of EBOV GP from different Ebolavirus strains. The sequences were ordered based on phylogenetic conservation (indicated by the phylogenetic tree). Protein backbones are depicted as broken gray lines, where spaces represent gaps in the alignment. Experimentally identified unambiguous O-glycosylation sites are marked as red circles. Conserved Ser/Thr amino acids are marked as yellow circles (including ±1 amino acids from a gap in the alignment), whereas partially conserved Ser/Thr (within ±3 amino acids from identified glycosite position) are marked as pink circles. (A) Full alignment. (B) Zoom in on the protein region containing the mucin-like domain.