Fig 2.
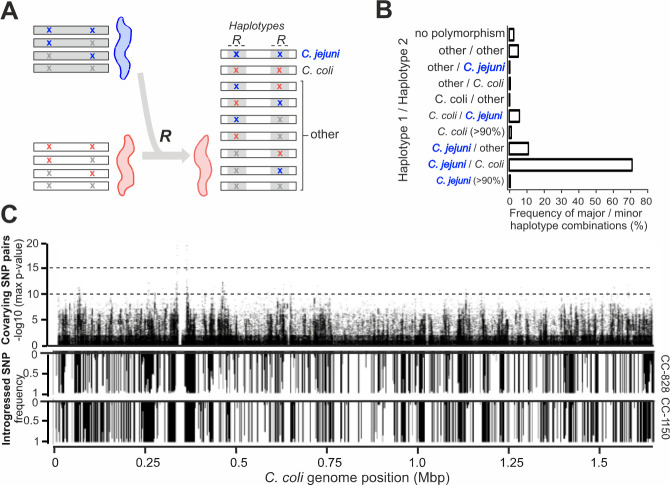
Covariation in introgressed C. coli genomes. (A) Possible SNP pairs in ancestral donor C. jejuni and recipient C. coli genomes and potential recombinant (R) haplotype pairs in introgressed C. coli. Possible genome haplotypes are designated with C. jejuni-fixed SNPs (blue X), other C. jejuni SNP (gray X), C. coli-fixed SNP (red X), and other C. coli SNPs (gray X). (B) The frequency of major and minor haplotype combinations among the 2,578 pairs of co-varying SNPs in the 689 C. coli clade-1-recipient genomes, revealing that 83.5% of long-range covariation was between introgressed C. jejuni sites. (C) Miami plot of each polymorphic site showing the maximum P-value for co-varying biallelic pairs (>20 kbp apart) and the frequency of introgression in CC-828 and CC-1150.