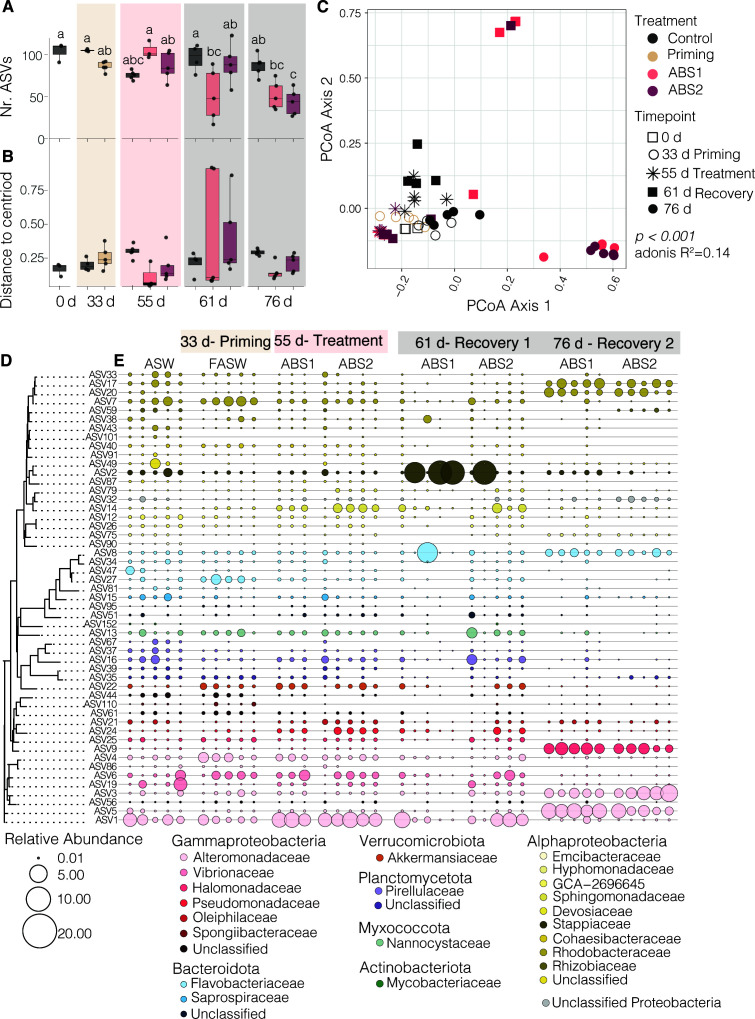
Fig 3.
Antibiotic exposure shifts the core microbiome of Aiptasia. (A) Boxplots showing alpha diversity, as reported as the number of observed ASVs, revealed that anemones exposed to antibiotics had a significantly less diverse community in comparison to the controls (n = 4–5 per treatment per time point)(two-way ANOVA P < 0.001). (B) Boxplots of beta-dispersion values and (C) a PCoA analysis calculated from a Bray-Curtis distance matrix of the core (D) microbiome revealed that beta-diversity significantly differs between treatment groups and across sampling time points (ANOSIMS R2 = 0.14, P < 0.004). In panel A, the letter represents results from a Tukey’s Honest Significant Difference test, where groups that are connected by the sample letter are not statistically different. (D) A dendrogram of the core microbiome based on sequence distances grouped taxa according to phylogenetic grouping. (E) Bubble plots showing shifts in the relative abundance of the core members across treatment groups and sampling time points. Only a subset of the ASW control samples collected at each sampling time point are displayed. All treatment samples are ordered by sampling time point and treatment conditions. The size of the bubble corresponds to the relative abundance of the ASV. If there is no point shown in the graph, the ASV was not detected in the sample. ASW = artificial seawater, FASW = filtered artificial seawater, ABS = antibiotic solution. Results of statistical tests are reported in Table S1 and the core community is identified in Table S3.