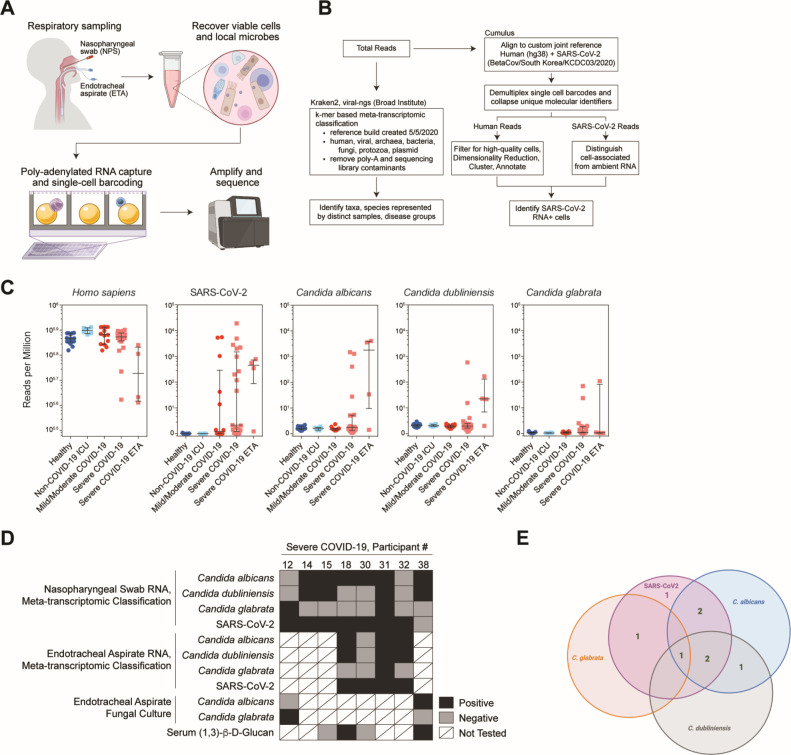
Fig 1.
Codetection of host single-cell transcriptome with intracellular and microenvironmental pathogen-derived genomic material. (A) Schematic of biological sample processing pipeline. (B) Schematic of computational pipeline for each sample. (C) Abundance of human, SARS-CoV-2, and Candida spp. by participant and disease group as defined by meta-transcriptomic classification. N = 56 participants. Lines represent median ±interquartile range. (D) Summary of results from sequencing, fungal culture, and serum (1,3)-β-D-glucan assays from eight participants with detectable Candida species reads by nasopharyngeal swab or endotracheal aspirate. (E) Graphical summary of COVID-19 and Candida sp. infection among severe COVID-19 cohort.