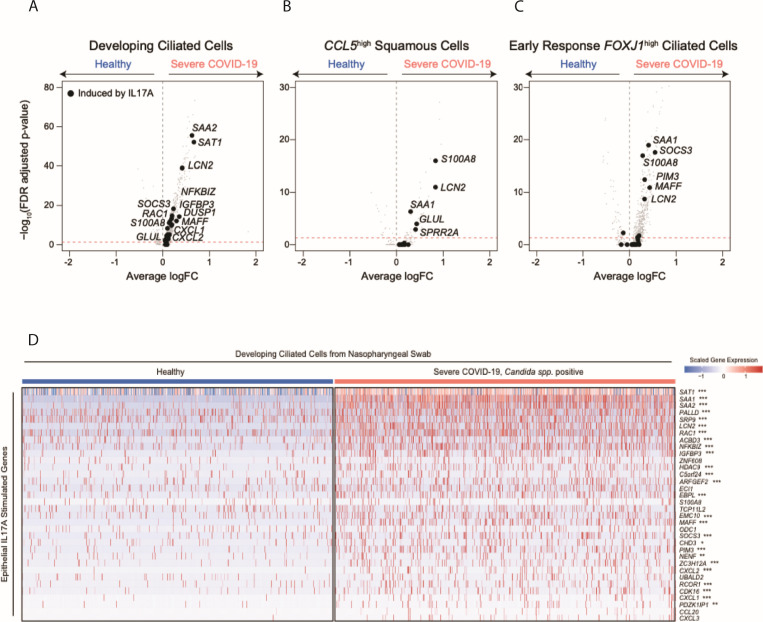
Fig 4.
Transcriptional responses of respiratory epithelial cells following in vitro exposure to various cytokines (A–C). Volcano plots of differentially expressed genes between select epithelial cell types from healthy participants vs those with severe COVID-19: developing ciliated cells (A), CCL5high squamous cells (B), and early response FOXJ1high ciliated cells (C). Gray points, all genes; black points, genes induced in human nasal epithelial cells following IL17A treatment (as in Fig. 3A). (D) Heatmap of IL17A-induced genes among developing ciliated cells from NP swabs. Heatmap colors reflect scaled gene expression: red, higher expression; blue, lower expression. Left columns (blue bar), developing ciliated cells from n = 15 healthy participants; right (pink bar), developing ciliated cells from n = 8 participants with severe COVID-19 and Candida spp. detected. Genes (rows) selected represent genes significantly upregulated among cultured human nasal epithelia cells following in vitro exposure to IL17A. Statistical significance comparing healthy-derived vs severe COVID-19, Candida spp. positive-derived single cells by likelihood ratio test assuming an underlying negative binomial distribution. ***FDR-corrected P < 0.001; **FDR < 0.01; *FDR < 0.05.