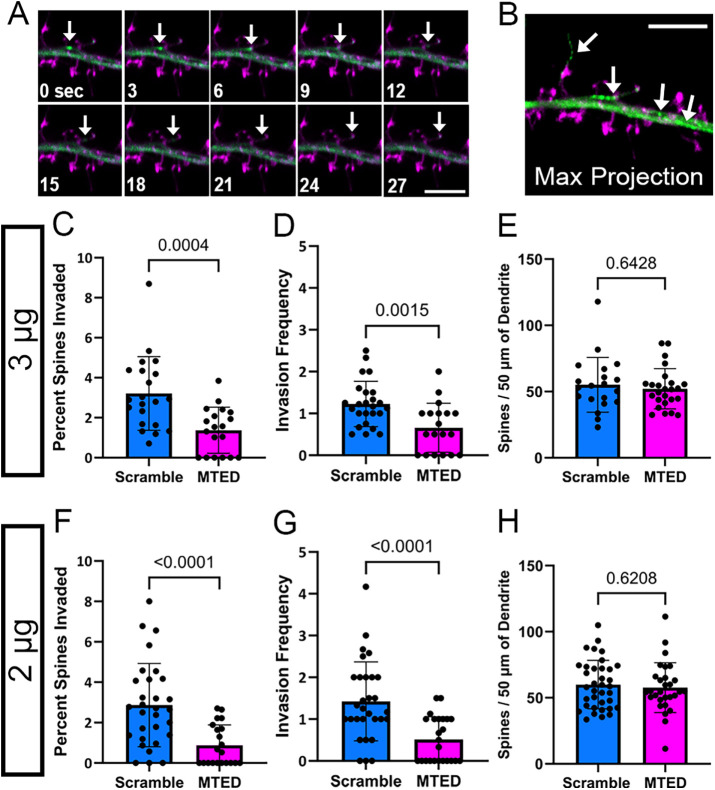
FIGURE 2:
Reduction of MT invasions into dendritic spines within LifeAct-MTED transfected hippocampal neurons. (A) Confocal time-series of living mature hippocampal neuron (DIV 23) taken at 3-s intervals. The neuron was transfected with LifeAct-scramble (magenta) and mNeon EB3 (green) plasmid constructs. Invading EB3 comet is shown with a white arrow in each frame. (B) Corresponding maximum intensity projection of the time-series shown in A. Traces of an invading EB3 comet, as well as EB3 comets within the dendritic shaft, are shown in green and are identified by white arrows. Dendritic spines of the transfected neuron are shown in magenta. Scale bars are 5 µm (A and B). (C–H) Bar graphs show mean ± SD and black dots are individual dendritic segments. (C) Quantification of percent of dendritic spines invaded (number of invaded dendritic spines/total number of dendritic spines within field of view) for neurons transfected with 3 μg of LifeAct-scramble or LifeAct-MTED plasmid (n = 21 scramble, n = 19 MTED from five separate biological replicates for both C and D). (D) Quantification of invasion frequency (total number of invasions throughout the course of a time-series containing 100 frames/invaded dendritic spines within field of view) for neurons transfected with 3 μg of LifeAct-scramble or LifeAct-MTED plasmid. (E) Quantification of dendritic spine density normalized to 50 µm dendritic segments for neurons transfected with 3 μg of LifeAct-scramble or LifeAct-MTED (n = 20 scramble, n = 25 MTED from six separate biological replicates). (F) Quantification of percent of dendritic spines invaded for neurons transfected with 2 μg of LifeAct-scramble or LifeAct-MTED (n = 30 scramble, n = 21 MTED from five separate biological replicates for both F and G). (G) Quantification of invasion frequency for neurons transfected with 2 μg of LifeAct-scramble or LifeAct-MTED plasmid. (H) Quantification of dendritic spine density normalized to 50 µm dendritic segments for neurons transfected with 2 μg of LifeAct-scramble or LifeAct-MTED (n = 36 scramble, n = 29 MTED from six separate biological replicates). P values in (C–H) shown above bars are calculated with two-tailed Student’s t test or Mann-Whitney depending on normality of data distribution.