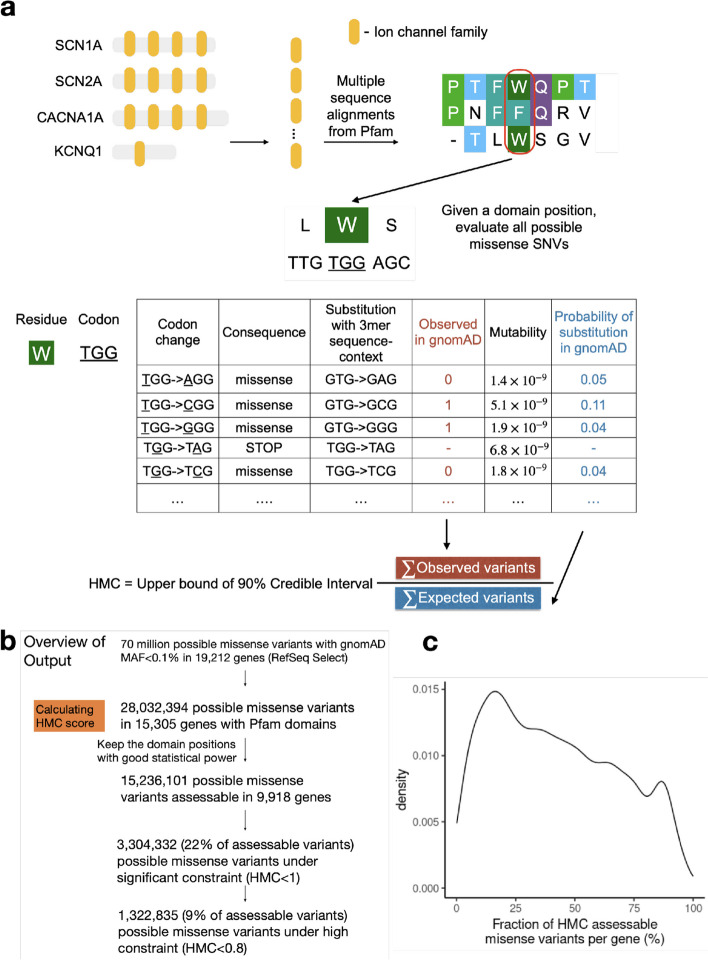
Fig. 1.
Overview of developing Homologous Missense Constraint. a Here, we illustrate how to calculate HMC scores using genes with ion channel domains (Pfam ID: PF00520). Evolutionarily equivalent residues were identified by aligning protein sequences across the protein domain family. Given a domain position with equivalent residues, the observed/expected ratio is calculated to measure the genetic constraint of missense variants at this domain position. HMC score is defined as the upper bound of 90% credible interval of the observed/expected ratio. HMC < 1 indicates significant (P-value < 0.05) depletion of missense variation at that residue, and missense variants at these positions are predicted as deleterious. b Summary of total number of missense variants evaluated by HMC. c Fraction of HMC assessable missense variants across genes (only include 9918 genes with assessable variants). For a few genes, nearly all missense variants are assessable while a few genes have very few assessable variants. For most genes, typically 20–63% variants are assessable (median = 40%)