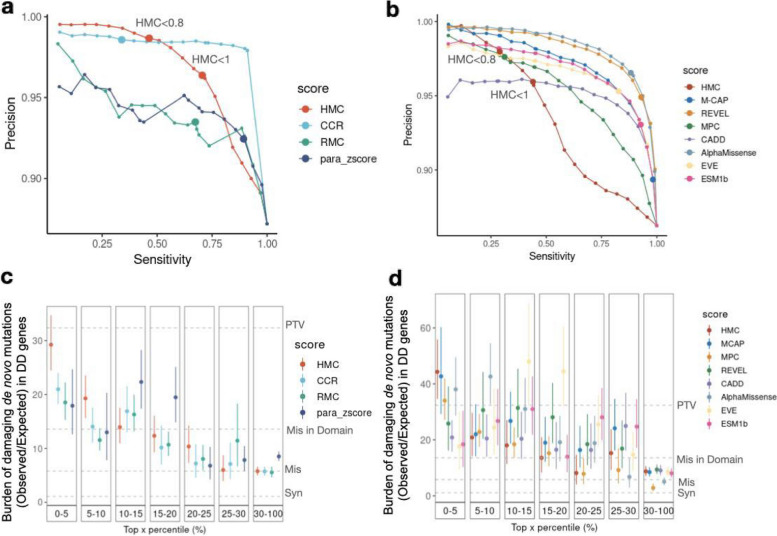
Fig. 3.
HMC has greater precision than other constraint metrics, and comparable performance to meta-predictor pathogenicity scores. a Using ClinVar variants, the precision-sensitivity curve demonstrates that HMC has higher precision over the constraint- and homologous-residue-based methods in top-ranked variants and within authors’ recommended thresholds (dots with larger size). The recommended threshold and the corresponding precision and sensitivity for each tool are HMC < 0.8 (98.6%, 48.8%), CCR > 95 (98.6%, 34%), RMC < 0.6 (93.5%, 67.4%) and para_zscore > 0 (92.4%, 89.3%). b HMC has comparable precision as existing state-of-the-art supervised meta-predictors. Dots with larger size indicate the performances (precision, sensitivity) using authors’ recommended thresholds: HMC < 0.8 (98.0%, 29.0%), M-CAP ≥ 0.025 (89.4%, 98.2%), REVEL > 0.5 (94.9%, 92.9%), CADD ≥ 10 (no variants are scored as deleterious), MPC ≥ 2 (97.6%, 31.2%), AlphaMissense ≥ 0.5 (96.5%, 88.3%), EVE ≥ 0.5 (95.3%, 82.8%), ESM1b ≤ − 7.5 (93.1%, 92.8%). c–d Using the 31 K trio data of DD, the burden of de novo mutations (observed/expected) in DD-associated genes is compared to evaluate the precision of predicting damaging variants (the higher the burden, the more likely the variants are damaging). For a given tool, variants are grouped into bins based on their percentile of predicted pathogenicity among all assessed variants. As baseline references, the dotted lines show the burden ratio for synonymous DNMs, missense DNMs, missense DNMs within annotated Pfam domains, and protein-truncating DNMs as shown in Fig. 2d. c Comparison between HMC with existing constraint-based and homologous residue-based scores using DNM burden and its 95% confidence interval; d Comparison between HMC with existing state-of-the-art meta-learners using DNM burden and its 95% confidence interval