Extended Data Figure 2. Upregulation of the mevalonate/cholesterol synthesis pathway in mouse and human TRM.
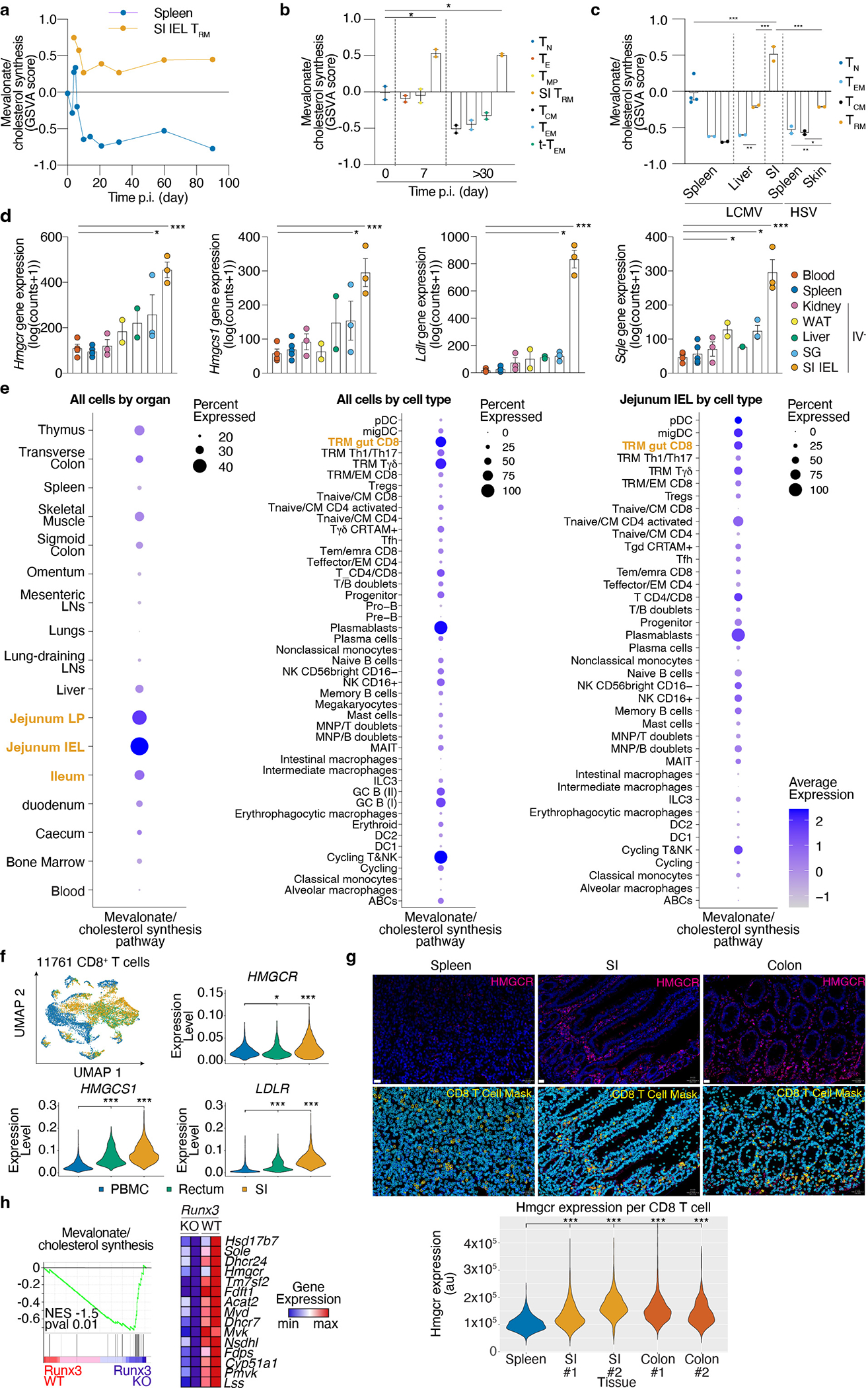
a, Mevalonate/cholesterol synthesis GSVA scores from averaged single cell expression of spleen and SI P14 at indicated time points after LCMV infection profiled by scRNASeq, GSE131847. b, Mevalonate/cholesterol synthesis GSVA scores for indicated subsets of CD8 T cells in the spleen or SI in the context of LCMV infection, GSE157072. c, Mevalonate/cholesterol synthesis GSVA scores for indicated subsets of CD8 T cells at the indicated tissues in the context of LCMV and Herpes Simplex Virus (HSV), GSE70813. d, Gene expression of indicated genes in P14 CD8 T cell populations from indicated tissues >30 days after LCMV infection, GSE182276 e, Mevalonate/cholesterol synthesis pathway scores for all human immune cells grouped by indicated tissues (left), cell types (middle), or cell types of the Jejunum SI (right), from the tissue immune cell scRNAseq atlas. f, Expression of selected genes on human CD8 T cells from PBMC, rectum, and SI samples, pooled from 13 healthy donors, GSE125527. g, HMGCR protein expression in CD8 T cells from human spleen, SI, and colon by immunofluorescence staining. Quantification of total HMGCR by CD8 T cell area. Donor ID (#). Scale bar, 20 μm. h, GSEA of the mevalonate/cholesterol synthesis in Runx3 KO or control CD8 T cells GSE106107. Data are pseudo-bulk averaged values (a), mean +/− s.e.m. (b, c, and d), or geometric distribution (f and g), with a total of n=2 (b), n=2–4 (c), n=2–5 (d) mice, and n=1–2 (g) samples. Two-sided unpaired t-Test (b, c, d, f, and g), and Gene Set Enrichment Analysis (h). *P<0.05, **P<0.01, ***P< 0.005.
