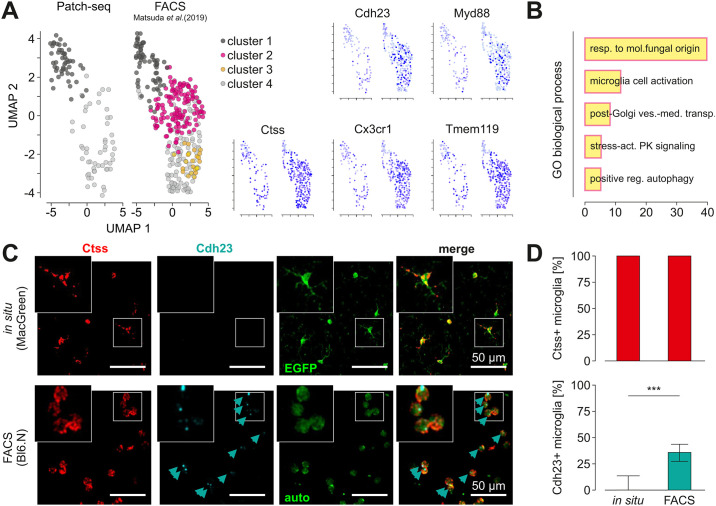
Fig 4. Comparison of single cell transcriptomes obtained by patch seq and after FACS sorting.
A Left, Comparison of microglia single cell data from the current patch-seq and a previously published data set after FACS isolation (Masuda et al., 2019). The latter data were obtained from microglia after brain dissociation and FACS. Note that clusters 1 and 4 appear in both data sets whereas clusters 2 and 3 are specific for FACS samples. Right, Gene expression analysis of the data sets shown in A for the microglia activation markers Cdh23, Myd88 as well as for the homeostatic genes Ctss, Cx3cr1 and Tmem119. B Gene ontology (GO) enrichment analysis of 275 top marker-genes of a FACS specific Cluster 2 reveals Cluster specific pathways. The bar chart represents the top 5 significantly enriched pathways in Cluster 2 in comparison to all other clusters. X-axis shows the fold enrichment of each pathway. C Detection of Ctss and Cdh23 in cortical brain slices from MacGreen mice (top) and FACS-isolated microglia from Bl6 mice (bottom) by RNAscope and confocal microscopy. Ctss (left) and Cdh23 (middle left) are indicated in red and cyan, respectively. Intrinsic EGFP signal of cell in slices is given in green (middle right); FACS isolated microglia in lower image show autofluorescence (auto). Merged images (right) show overlap of signals, indicated by arrows. The small square in each image is shown enlarged in the insets on the top left of each image. Scale bars: 50 μm. D Quantification of Ctss and Cdh23 in microglial cells in situ and after FACS isolation. Note that the homeostatic microglial marker Ctss (left) was expressed in both, in situ and in vitro microglial cells whereas Cdh23 (middle) was significantly more present after FACS sorting. Significane was tested by Mann-Whitney (p < 0.0001).