Extended Data Fig. 10. Validation of pharmacological strategies to overcome intratumor heterogeneity in high-grade meningiomas.
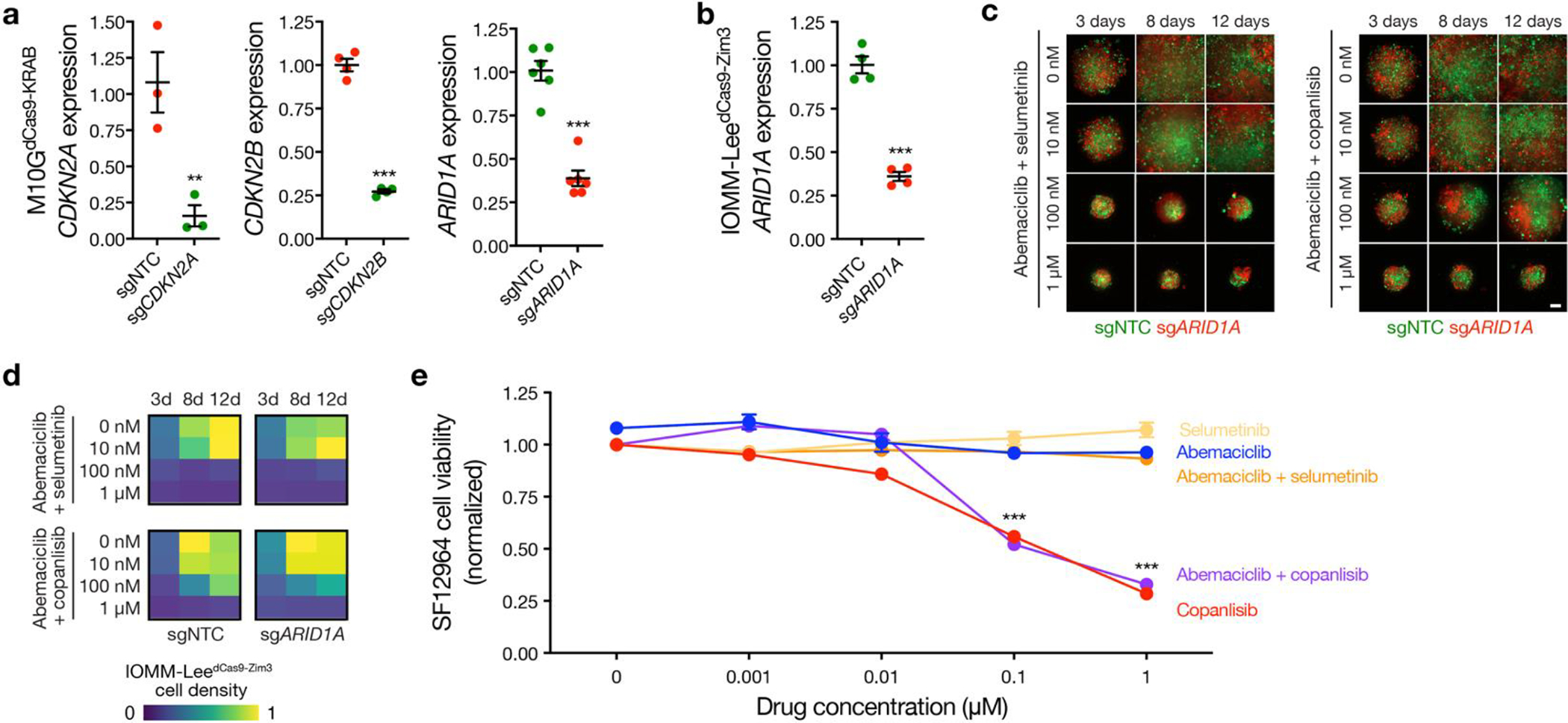
a, M10G patient-derived meningioma cells stably expressing CRISPRi machinery (dCas9-KRAB) and sgRNAs suppressing CDKN2A (sgCDKN2A, n=3), CDKN2B (sgCDKN2B, n=4), ARID1A (sgARID1A, n=6), or non-targeted control sgRNAs (sgNTC, n=3, 4, 6, respectively). Cells were labeled with red or green fluorescence proteins and integrated into 3D co-cultures for pharmacologic and live cell imaging experiments. b, IOMM-Lee meningioma cells that lack endogenous CDKN2A/B stably expressing CRISPRi machinery (dCas9-Zim3) and sgARID1A (n=4) or sgNTC (n=4). Cells were labeled with red or green fluorescence proteins and integrated into 3D co-cultures for pharmacologic and live cell imaging experiments. c, Combination molecular therapy treatments of 3D co-cultures of IOMM-LeedCas9-Zim3 meningioma cells expressing sgARID1A or sgNTC. Scale bar, 100μm. d, Quantification of combination molecular therapy treatments of 3D co-cultures of IOMM-LeedCas9-Zim3 meningioma cells expressing sgARID1A or sgNTC. Representative of 8 biological replicates per condition. e, MTT cell viability results normalized to vehicle control after 5 days of molecular therapy of SF12964, a patient-derived meningioma cell line from a WHO grade 3 meningioma (Hypermitotic DNA methylation group, NF2 p.Q165* mutation, chromosome 1p and 22q deletion, CDKN2A/B homozygous deletion, chromosome 1q amplification) that underwent reoperation after 5 months of abemaciclib treatment following prior surgeries and prior radiotherapy treatments. Representative of 8 biological replicates per condition. Lines represent means and error bars represent standard error of the means. Student’s t tests, one-sided, **p≤0.01, ***p≤0.0001.
