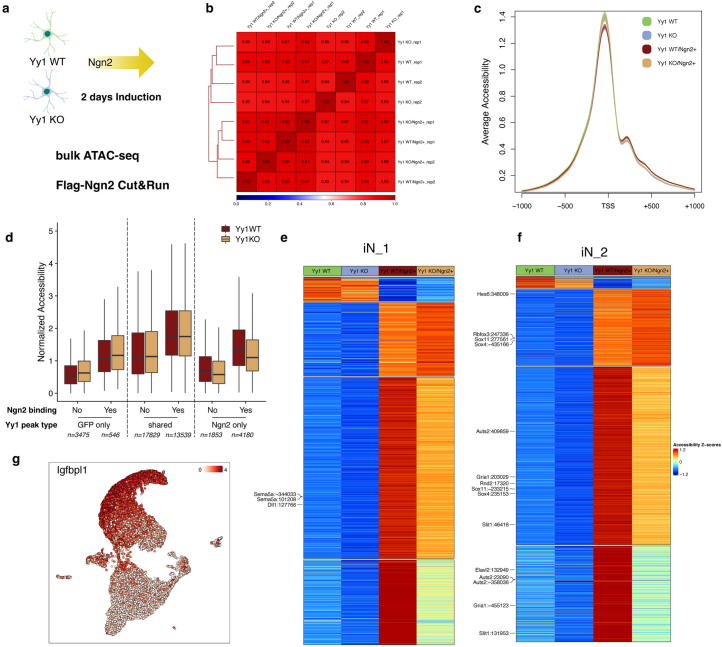
Extended Data Fig. 7. Yy1 deletion impairs Ngn2-mediated chromatin remodelling.
(a) Schematic representation of the experimental setup (b) Pearson’s correlation for ATAC in the indicated experimental conditions (N = 2 biological replicates). (c) Average accessibility (+/- 1 kb) at all TSSs in the indicated experimental conditions. (d) Boxplots depicting the normalized accessibility levels in Yy1WT/KO + Ngn2 conditions at different categories of Yy1 peaks (based on Fig. 7a) grouped by the overlap with Ngn2 binding sites. Boxplots display median (line), 25th or 75th percentiles (box),10th or 90th percentiles (whisker) and outliers (dots) (e-f) Heatmaps depicting the accessibility levels at iN_1 or iN_2 distal enhancers in the indicated experimental conditions. (g) UMAP visualization of the expression levels of Igfbpl1.