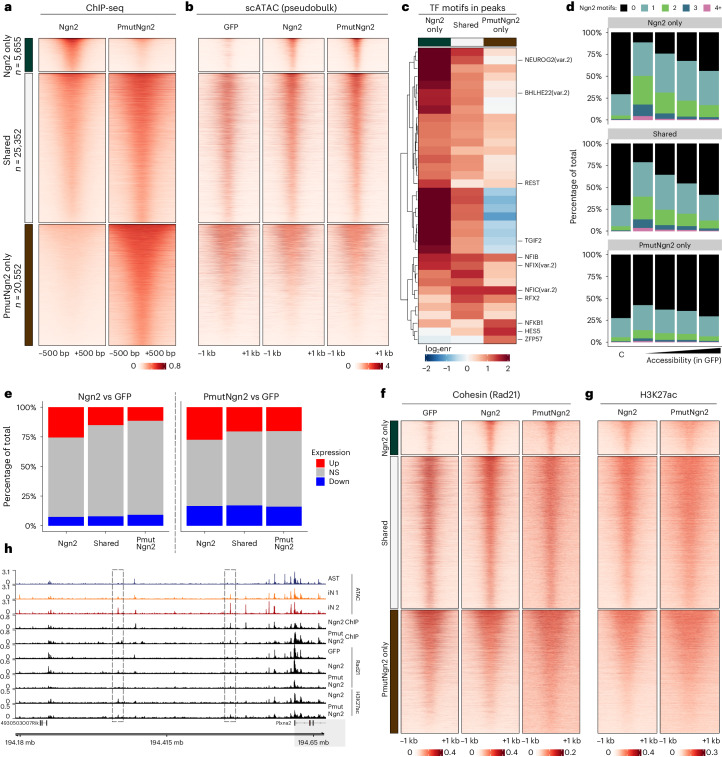
Fig. 4. Ngn2 or PmutNgn2 differentially bind and remodel chromatin.
a,b, Heatmaps showing the enrichment of ChIP-seq (a) or pseudobulk single-cell ATAC-seq (b) signal around differentially bound or shared peaks in murine iNs. c, Heatmaps depicting the motif enrichment in the peaks groups shown in a and b. d, Number of Ngn2 motifs in different peak categories, stratified by chromatin accessibility in GFP-transduced astrocytes. ‘C’ indicates the control group of peaks (randomly sampled accessible regions that are not bound by Ngn2 or PmutNgn2). e, Percentage of differentially regulated genes (based on bulk RNA-seq) overlapping with different peak categories (±100-kb window around the TSS). f,g, Heatmaps showing the enrichment of cohesin (Rad21) (f) or H3K27ac (g) CUT&RUN signal around differentially bound or shared Ngn2/PmutNgn2 peaks. h, Genome track showing aggregated single-cell ATAC accessibility, Ngn2/PmutNgn2 ChIP-seq as well as Rad21 and H3K27ac CUT&RUN tracks in the indicated experimental conditions at the Plxna2 locus. Values represent reads per million mapped reads (RPM). Dashed rectangles depict distal enhancers of the Plxna2 locus (shaded region), which show increased chromatin accessibility and recruitment of Rad21 and H3K27ac upon Ngn2 binding. enr, enrichment; NS, not significant.