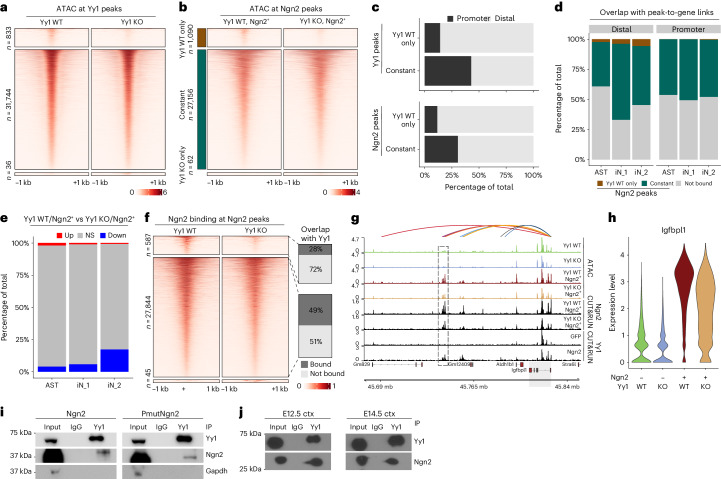
Fig. 8. Yy1 facilitates Ngn2-mediated epigenetic remodeling.
a, Heatmaps showing the enrichment of chromatin accessibility signal around differentially bound or shared Yy1 peaks in astrocytes from Yy1 WT or Yy1 KO mice, respectively. b, Heatmaps showing the enrichment of chromatin accessibility signal around differentially accessible or shared Ngn2 peaks in either Yy1 WT+Ngn2 or Yy1 KO+Ngn2 condition. c, Overlap of Yy1 and Ngn2 peaks that lose accessibility (Yy1 WT) or remain unchanged (shared) with genomic features. d, Overlap of Yy1 and Ngn2 peaks that lose accessibility or remain unchanged with EGP links as identified in Fig. 3g. e, Percentage of differentially regulated genes (based on single-cell RNA-seq) overlapping with either distal or promoter EGP anchor. f, Heatmaps showing Ngn2 (Flag) CUT&RUN enrichment at either differentially bound or shared peaks. Shown is also the ratio between peaks overlapping with Yy1 in each category. g, Genome track showing the linked enhancers (arcs), chromatin accessibility, Ngn2 and Yy1 CUT&RUN signal in the indicated conditions at the Igfbpl1 locus. A cluster of enhancers with reduced accessibility upon Yy1 KO is indicated with a dashed rectangle. The shaded region highlights the Igfbpl1 locus. h, Violin plot depicting the expression levels of Igfbpl1 in the indicated conditions. i, Co-IP experiments in P19 cells showing pulldown using IgG or Yy1 antibody and staining for Yy1, Ngn2 and Gapdh. j, Similar to i but using either E12.5 or E14.5 cortex with endogenous Yy1 and Ngn2. i,j, Co-IP experiments were independently replicated (n = 2). ctx, cortex; KO, knockout; NS, not significant; WT, wild-type.