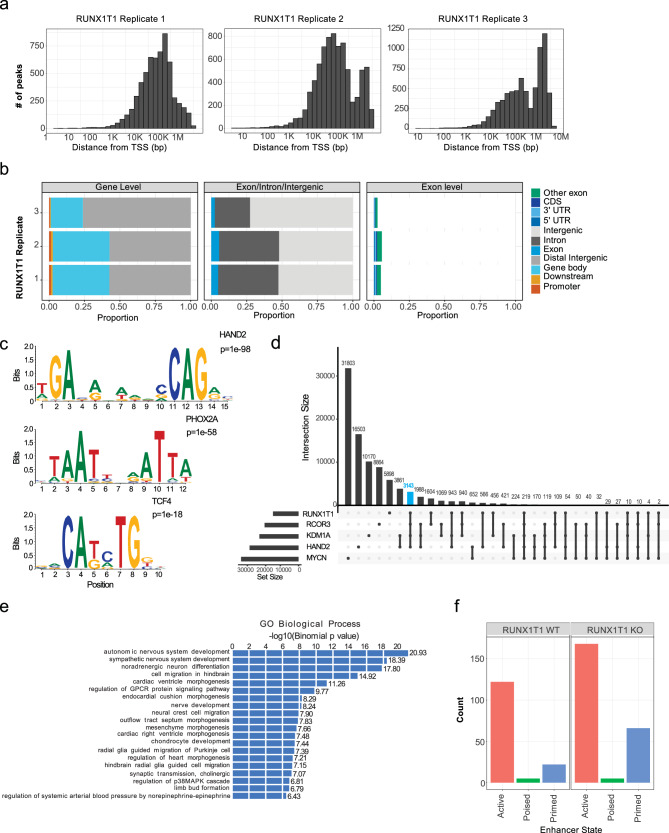
Fig. 7. RUNX1T1 binds to intergenic regions and interacts with HAND2 to maintain an undifferentiated neuroblastoma phenotype.
a ChIP-seq analysis of RUNX1T1 binding across three replicates showing the number of peaks and their distance in base-pairs (bp) to the transcription start site (TSS). b Proportion of the peaks occurring at different binding site locations from the RUNX1T1 three ChIP-seq replicates. RUNX1T1 binding occurring at either a promoter, gene body, distal intergenic or downstream in relation to a gene (left), binding in an exon, intron or intergenic (middle), and binding within exons at either a coding region (CDS), 5’ untranslated region (UTR) or 3’ UTR (right). c Motif discovery using homer analysis showing significantly enriched motifs from HAND2, PHOX2A, and TCF4. d UpSet plot of the intersection of ChIP-seq peaks of RUNX1T1, HAND2, LSD1, and RCOR3. The number of peaks identified are indicated for each gene and the intersection of gene peaks. RUNX1T1 peaks that co-localized with HAND2, LSD1, and CoREST3 are shown by the blue bar. e GREAT analysis (using Binomial test) showing enriched gene ontology (GO) biological process of the common peaks between RUNX1T1, HAND2, LSD1, and RCOR3. f Number of active, poised, and primed enhancer sites where RUNX1T1 binds in the presence (wildtype; WT) and absence of RUNX1T1 (knock out; KO).