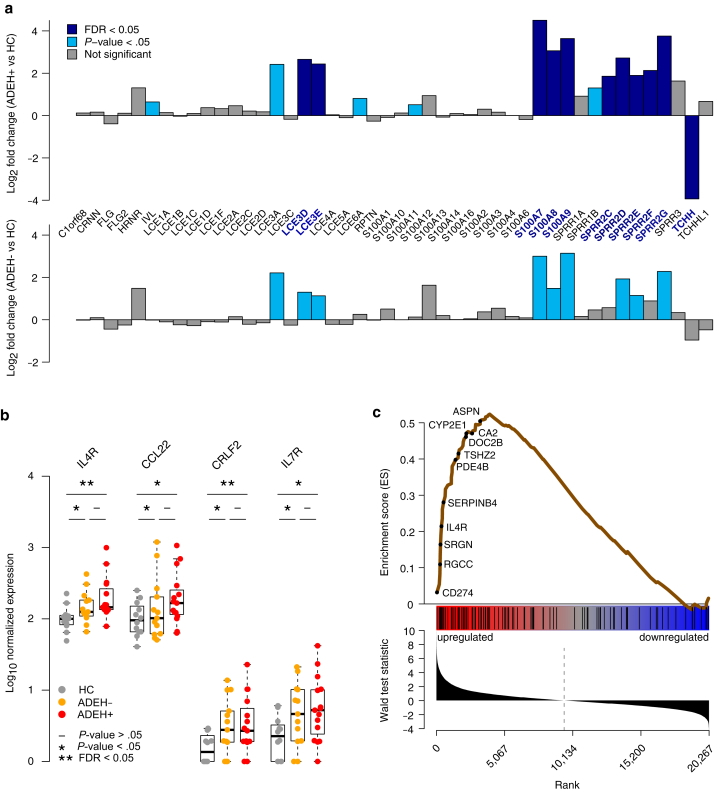
Figure 2.
Dysregulation of EDC gene expression in ADEH+ corresponds with T2 inflammation. (a) Bar plots of LFCs for EDC genes, comparing epidermal ADEH+ with HC skin (top) and ADEH− with HC skin (bottom). Teal = nominally significant differences (P < .05); dark blue = significant differences (FDR < 0.05). Many dysregulated EDC genes in ADEH+ skin are similarly dysregulated in ADEH− skin. (b) Box plots of epidermal expression for canonical T2 genes in HC (gray), ADEH− (orange), and ADEH+ (red) samples. Asterisk (∗) denotes nominally significant differences (P < .05); asterisks (∗∗) denote significant differences (FDR < 0.05). (c) ES plot from GSEA visualizing enrichment of ADEH+ epidermal DEGs within a gene set preranked on the basis of DE analysis between T2-high and T2-low AD. Top: running-sum ES profile (brown line) across T2-ranked genes, where the left-side peak of positive ES values indicates particular enrichment of ADEH+ DEGs among the most T2-upregulated genes. A subset of genes belonging to the leading edge of enrichment (appearing at/before the ES maximum and most strongly contributing to enrichment) are indicated. Middle: vertical black bars denote positions of ADEH+ DEGs along the T2-ranked list of genes. Bottom: bar plot showing the ordered distribution of the gene ranking statistic (Wald test statistic from DESeq2). AD, atopic dermatitis; ADEH, atopic dermatitis with eczema herpeticum; DE, differential expression; DEG, differentially expressed gene; EDC, epidermal differentiation complex; ES, enrichment score; FDR, false discovery rate; GSEA, gene set enrichment analysis; HC, healthy control; LFC, log2 fold change; T2, type 2 cytokine.