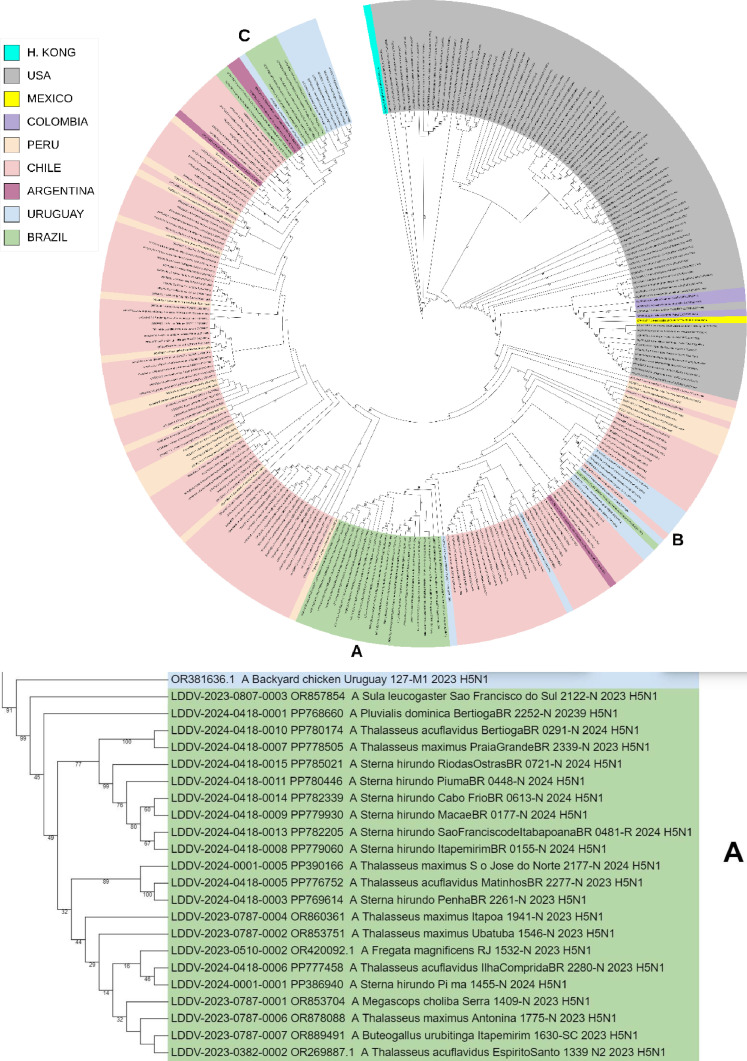
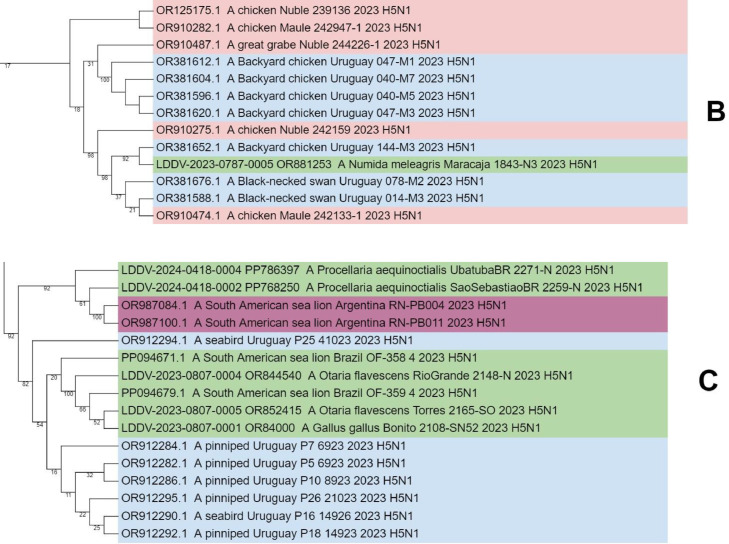
Fig. 1.
Phylogenetic analysis of highly pathogenic avian influenza A(H5N1) from wild birds, poultry, and mammals, Brazil. Maximum-likelihood method was used for phylogeny of hemagglutinin H5 sequences from avian influenza viruses in IQ-TREE multicore v.1.6.12. Phylogenetic tree was generated with Geneious Prime® 2024.0.3 software and edited with iTOL tool (https://itol.embl.de/). Sequences were aligned by using the MAFFT program in the Geneious Prime® 2024.0.3 software and editor for the same software. We used the best-fitted evolutionary model based on BIC = Bayesian Information Content models; robustness of tree topology was assessed with 1000 bootstrap replicates Green color indicate clustering of strains from Brazil and sequences from this study; “LDDV” indicates the sequences from this study. Non–goose/Guangdong lineage virus strains from Eurasia were outgroups. A, B and C shares a most recent common ancestor with viral lineages of Chile, Uruguay, and Argentina (2022–2024). With 82–92% bootstrap support values, confirming its closest phylogenetic relatedness with the sequences from South American.