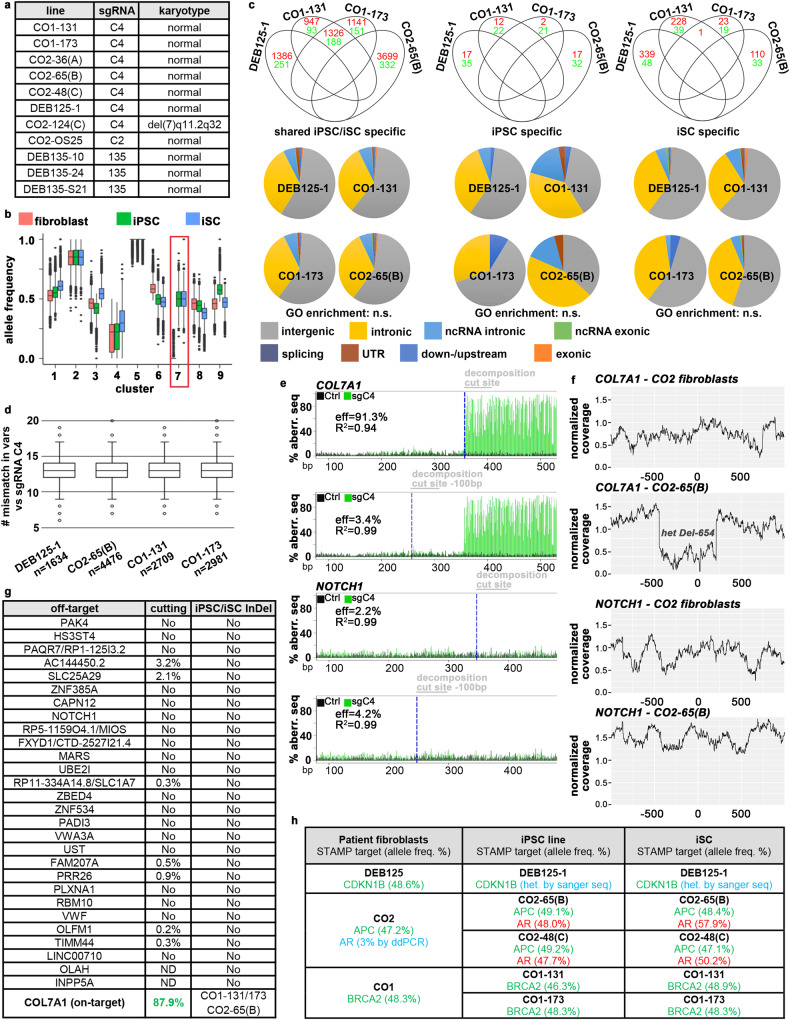
Fig. 4. Genomic and chromosomal stability.
a Normal karyotypes in 10 of 11 iPS cell lines from four patients. b k-means clustering of all variants (n = 111,741) found by whole-genome sequencing (WGS) in fibroblasts, iPS cells and iSCs from patient DEB125 (see Supplementary Data 2). Red frame highlights a sub-set with differential allele frequencies (AFs) in iPS cells/iSCs compared to fibroblasts. Boxes: interquartile ranges (25–75%), center lines: medians, whiskers: 1.5 times the interquartile range; outliers: circles. c Cutoff/odds ratio filtering (see Methods) identifies variants from WGS (red: SNPs; green: InDels) specifically found in cell types from indicated patient lines. Grouping in Venn diagrams indicates the absence of positive variant selection. The majority of cell-type-specific variants is found in intergenic or intronic sequences (pie charts). No gene ontology (GO) term enrichment detected. d Aligning all shared iPS cell/iSC-specific variants with the used seed sequence of sgRNA C4 within a 25 bp search window that must contain a NRG PAM-motif does not identify any homologies. The outlier (circles) with the highest similarity exhibits six mismatches. Boxes: interquartile ranges (25%–75%), center lines: medians, whiskers: 1.5 times the interquartile range. e TIDE analysis of sgRNA C4-mediated CAS9 cutting in CO2 fibroblasts at the COL7A1 on-target (top) and the in silico predicted off-target NOTCH1 (bottom). Measurements taken upstream (Ctrl) and downstream of cut sites. f Plots of normalized WGS coverage 1000 bp up-/downstream of COL7A1 (top) and NOTCH1 (bottom) from fibroblasts/iPS cells. Note the sharp drop of coverage at the COL7A1 locus in iPS cells due to a heterozygous 654 bp deletion (Fig. 2d–g). g Summary of all in silico predicted exonic and intronic off-targets as in (e, f). For TIDE (middle), controls were subtracted from cut sites. Heterozygous 1 bp, 10 bp, and 654 bp COL7A1-deletions were detected via WGS coverage (right; see Supplementary Data 3) in iPS cells/iSCs from homozygous patients. h Variants identified via the STAMPv2 oncopanel. Germline variants (green) are found in all 3 cell types. A heterozygous androgen receptor mutation (red) stems from clonal expansion of a fibroblast subpopulation (3%) with this lesion. Blue: variants identified by secondary methods. Source data are provided as a Source Data file.