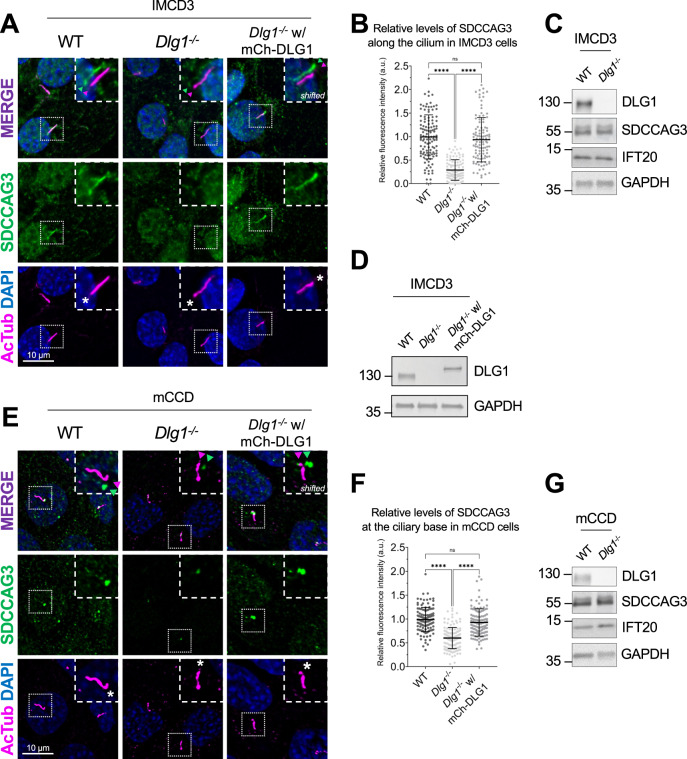
Figure 3. Loss of DLG1 impairs ciliary localization of SDCCAG3 in IMCD3 and mCCD cells.
(A) Immunostaining of ciliated cilia-BioID2 IMCD3 cell lines showing comparative SDCCAG3 staining (green) in WT, Dlg1−/− and mCherry-DLG1 (mCh-DLG1) rescue cells. Cilia were stained with antibodies against acetylated α-tubulin (AcTub, magenta), and nuclei visualized with DAPI staining (blue). Insets show enlarged images of cilia, and asterisks mark the ciliary base. The merged insets show primary cilia with channels shifted to aid visualization. (B) Quantification of the relative mean fluorescence intensity (MFI) of SDCCAG3 staining along the cilium of cilia-BioID2 IMCD3 cell lines. Graphs represent WT normalized and accumulated data of three independent biological replicates. The number of dots in each condition represents the total number of primary cilia quantified. Kruskal–Wallis test with Dunn’s multiple comparison test was used for the statistical analysis. Error bars represent mean ± SD. ****P < 0.0001; ns, not statistically significant. (C, D) Western blot analysis of total cell lysates of cilia-BioID2 IMCD3 cell lines. Blots were probed with antibodies as indicated, and GAPDH was used as a loading control. Molecular mass markers are shown in kDa to the left. (E) Immunostaining was done with anti-SDCCAG3 antibody (green) and anti-acetylated α-tubulin (AcTub, magenta) in ciliated mCCD WT, Dlg1−/− and mCherry-DLG1 (mCh-DLG1) rescue cells. Nuclei were visualized with DAPI staining (blue). Insets show enlarged images of cilia; asterisks mark the ciliary base. The merged insets show primary cilia with channels shifted to aid visualization. (F) Quantification of the relative MFI of SDCCAG3 staining at the ciliary base of mCCD cell lines. Graphs represent WT normalized and accumulated data of three independent biological replicates. The number of dots in each condition represents the total number of primary cilia quantified. Kruskal–Wallis test with Dunn’s multiple comparison test was used for the statistical analysis. Error bars represent mean ± SD. ****P < 0.0001; ns, not statistically significant. (G) Western blot analysis of total cell lysates of mCCD cell lines. Blots were probed with antibodies as indicated, and GAPDH was used as a loading control. Molecular mass markers are shown in kDa to the left. Source data are available online for this figure.