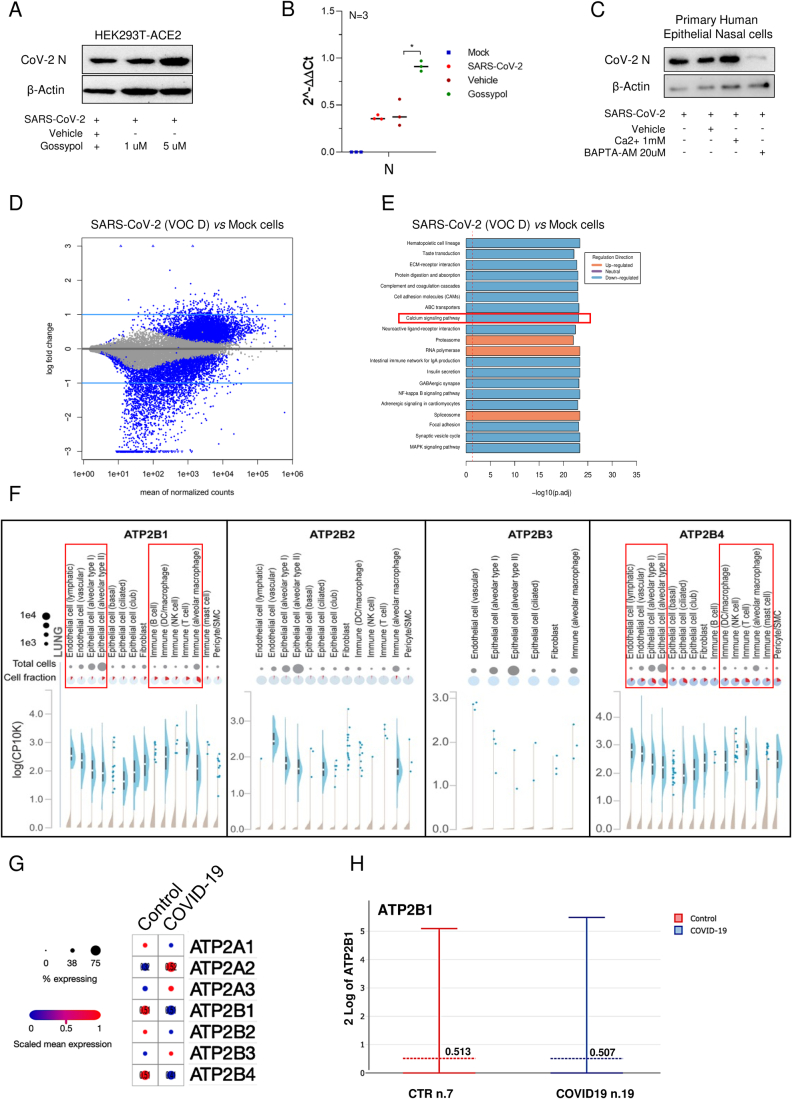
Figure EV1. Deregulated Ca2+ pumps during SARS-CoV-2 infection, including ATP2B1. Related to Fig. 1.
(A) A representative immunoblotting analysis using antibodies against the CoV-2 N protein from cells treated with escalating concentration of “gossypol-pubChem CID 3503” at 1 and 5 μM and vehicle as control in SARS-CoV-2 (VOC Δ) infected HEK293T-ACE2 cells. β‐Actin was used as the loading control. (B) Quantification of Cov-2 N gene (2−ΔΔCt) in gossypol-treated HEK293T-ACE2 (5 μM—48 h) and infected as in (A). Cells treated with vehicle were used as negative control. Scattered plot shows the individual value and mean as indicated by the horizontal black lines of N = 3 biological replicates. Unpaired two‐tailed T Student tests Bonferroni corrected. *p < 0.05 (vehicle vs. gossypol); the other comparisons are not statistically significant, as expected. (C) A representative immunoblotting analysis using antibodies against the Cov-2 N protein on total protein lysates obtained from human primary epithelial nasal cells treated with BAPTA at 20 μM concentration. Vehicle-treated cells were used as control. β‐Actin is used as the loading control. (D) RNA Sequencing (RNA-seq) analyses was performed in HEK293T-ACE2 cells treated as in Fig. 2E. (E) The Gene Set Enrichment Analysis (see GSEA project on https://doi.org/10.1073/pnas.0506580102) is applied for the identification of deregulated key genes and pathways. KEGG pathways analyses by statistical KS global test, P < 0.05. KEGG pathway enrichment analysis indicates those significant deregulated genes were highly clustered in calcium signaling pathway (red box). P adj: adjusted P values. (F) In silico analysis of publicly available datasets of single-cell RNA sequencing (https://singlecell.broadinstitute.org) for the expression of the plasma membrane calcium ATPases members (PMCAs or ATP2B1-4) of the large family of type Ca2+ion pumps in multiple cell type in the lung parenchyma (including alveolar macrophages and in the alveolar epithelial cells type I and type II). (G) Literature public search on available datasets obtained from a single-nuclei RNA-seq (snRNA-seq) on >116,000 nuclei from n.19 COVID-19 autopsy lungs and n.7 pre-pandemic controls (Melms et al, 2021); to verify expression of PMCAs and SERCAs pumps (ATP2B1-4 and ATP2A1-3 genes, respectively). The numbers within the dots are the median percentage level of expression between the two populations tested. (H) Expression levels of ATP2B1 in single-nuclei RNA-seq database (snRNA-seq) as described above in (G). Source data are available online for this figure.