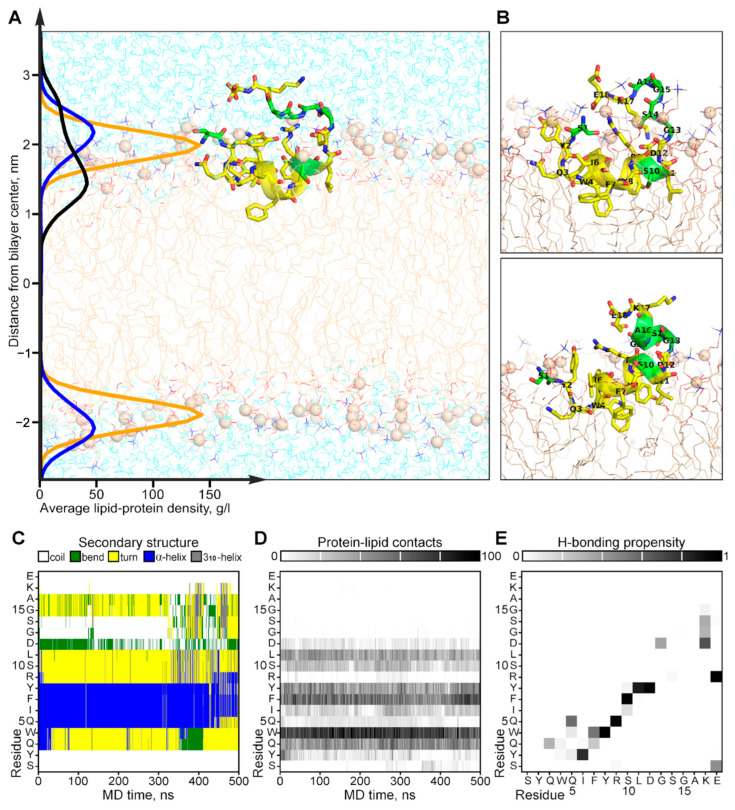
Figure 5.
Results of MD simulation of the NMR-derived structure of CpRE12 in POPC bilayer. (A) Representative MD snapshot with CpRE12 embedded into hydrated explicit POPC bilayer. The peptide is given in ribbon presentation, glycine, alanine and serine residues are shown in green. Phosphorus atoms of the lipid headgroups are shown by orange spheres. The density distributions of the peptide (in black), phosphorous (in yellow) and choline (in blue) groups of lipids, averaged over MD trajectory, are presented on the left. (B) Alternative conformations of CpRE12 observed in MD simulation. (C,D) Color-coded representation of the MD time evolution of the secondary structure and protein-lipid contacts of CpRE12 embedded into POPC bilayer. The secondary structure elements are shown in blue—α-helix, in gray—310-helix, in yellow—turn, in green—bend, in white—coil. Protein-lipid contacts are color-coded according to the number of direct van der Waals contacts between atoms with 5 Å distance cut-off from white (0 contacts) to black (100 protein-lipid contacts). (E) Propensity of H-bond formation between all backbone and side chain atoms of CpRE12, estimated over the MD trajectory.