Abstract
Alphavirus replicon vectors are well suited for applications where transient, high-level expression of a heterologous gene is required. Replicon vector expression in cells leads to inhibition of host macromolecular synthesis, culminating in eventual cell death by an apoptotic mechanism. For many applications, including gene expression studies in cultured cells, a longer duration of transgene expression without resulting cytopathic effects is useful. Recently, noncytopathic Sindbis virus (SIN) variants were isolated in BHK cells, and the mutations responsible were mapped to the protease domain of nonstructural protein 2 (nsP2). We report here the isolation of additional variants of both SIN and Semliki Forest virus (SFV) replicons encoding the neomycin resistance gene that can establish persistent replication in BHK cells. The SIN and SFV variant replicons resulted from previously undescribed mutations within one of three discrete regions of the nsP2 gene. Differences among the panel of variants were observed in processing of the nonstructural polyprotein and in the ratios of subgenomic to genomic RNAs. Importantly, high-level expression of a heterologous gene was retained with most replicons. Finally, in contrast to previous studies, efficient packaging was obtained with several of the variant replicons. This work expands the utility of noncytopathic replicons and the understanding of how alphavirus replicons establish persistent replication in cultured cells.
Alphavirus vectors, derived principally from Sindbis virus (SIN), Semliki Forest virus (SFV), and Venezuelan equine encephalitis virus, are widely used for gene expression studies in vitro and are being developed for both vaccine and gene therapy applications (25). Typically, these vectors are constructed in a format known as a replicon, due to the self-amplifying nature of the vector RNA (30). Replicons contain both the cis and trans alphavirus genetic elements required for RNA replication, as well as heterologous gene expression via the native subgenomic promoter. Upon introduction into cells, replicon RNA is translated to produce four nonstructural proteins (nsPs), which together comprise the alphaviral replicase. Replication proceeds through a minus-strand RNA intermediate and subsequently generates two distinct positive-strand RNA species, corresponding to a genomic-length vector RNA and an abundant subgenomic RNA encoding the heterologous gene (27). The replicon RNA can be packaged into virion-like particles by providing the structural proteins in trans, from in vitro-transcribed defective helper RNA (4, 15–17) or using packaging cell lines (16). Alternatively, the replicon RNA can be introduced directly into cells as plasmid DNA (2, 6, 8, 13).
In most mammalian cells, host macromolecular synthesis is inhibited following the introduction of alphavirus replicons, leading to eventual cell death by an apoptotic mechanism (11, 25). Thus, application of these vectors for some gene therapy applications and extended gene expression studies in cultured cells is limited. Given the many other attractive features of the alphavirus replicon system, it would be useful to extend the utility of these vectors to include long-term expression and reduced cytopathogenicity options.
Under appropriate conditions, alphaviruses and alphavirus-derived vectors can establish persistence in cultured cells (14, 26, 29) or exhibit delayed onset of cytopathic effects (9). The establishment of SIN replicon persistence in BHK cells has been associated with mutation of the protease domain of nsP2 (7, 10), and studies have suggested that the use of such mutants for long-term expression may be possible (1, 3). It remains to be determined whether mutation of other alphavirus nsPs or nsP2 domains can provide a noncytopathic phenotype by a similar or alternative mechanism.
To expand the utility of the noncytopathic replicon and further explore how persistence is established, we isolated additional SIN replicons with this phenotype, as well as SFV replicons with a similar phenotype. Mutations that conferred the establishment of persistent replication were mapped to several regions of nsP2 for both SIN and SFV replicons, in addition to the same residue 726 mutation identified previously (7, 10). These mutations had various effects on the levels of genomic and subgenomic replicon RNA and, in some cases, processing of the nonstructural polyprotein.
Selection of replicons that establish persistent replication.
To select alphavirus replicon variants capable of establishing persistent replication, the neomycin phosphotransferase gene (neo) was placed under the control of the subgenomic promoter in both SIN- and SFV-derived replicons. These plasmids, designated pSINBV-neo and pSFV-neo, were derived from pRSIN (8) and pSFV1 (15) (GIBCO-BRL), respectively. neo-containing replicons were transcribed in vitro from linearized plasmid template; in some experiments, the DNA templates were subjected to prior random mutagenesis using the bacterial strain XL-1 Red Mutator (Stratagene). Replicon RNAs were transfected into BHK cells, and the cells were subjected to G418 selection (Geneticin; 0.5 mg/ml; GIBCO-BRL) at 24 h posttransfection. Drug-resistant colonies were obtained from both nonmutagenized and mutagenized replicons. In addition, colonies were obtained after infection of BHK cells with packaged vector particles containing nonmutagenized neo replicon, generated as previously described (16). These data indicated that drug resistance was associated with the replicon RNA and that adaptive replicon mutations could occur within the cell.
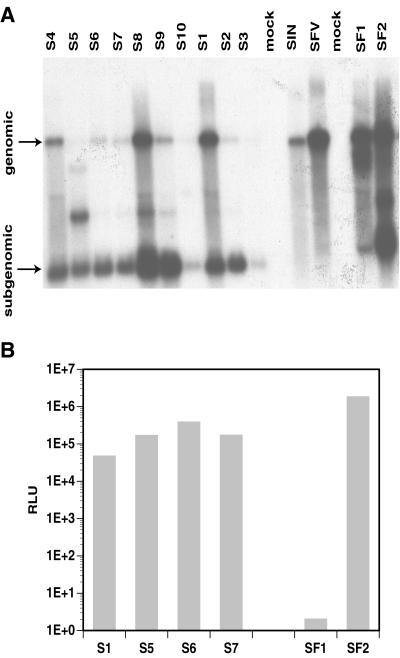
Within each selection, the drug-resistant BHK cells were pooled and expanded. To confirm that neo expression was associated with alphavirus replicon RNA species, poly(A)-selected mRNA was extracted from the pools (Triazol [GIBCO-BRL] followed by Oligotex [Qiagen]) and analyzed by Northern blot hybridization with a 32P-labeled DNA fragment derived from the neo gene (Fig. 1A). The neo sequence indeed was found within both genomic- and subgenomic-length RNA species for all pools. This analysis also indicated variations in the RNA profiles among SIN and SFV pools, particularly with respect to the relative ratios between subgenomic and genomic RNA and the appearance of new RNA species migrating faster than the genomic RNA (Fig. 1A, lanes S5, S8, S9, SF1, and SF2). Such variation suggested possible phenotypic differences among the selected variants.
FIG. 1.
Analysis of Neo-resistant BHK pools derived from SIN and SFV replicons. The SIN-derived pools are designated S1 to S10; the SFV-derived pools are named SF1 and SF2. (A) Northern blot analysis of poly(A)-selected RNA extracted from BHK cells either transfected (lanes S1, S2, S4 to S10, SF1, and SF2) or infected (lane S3) with replicon RNA and selected with G418. Pools were obtained using nonmutagenized replicon (lanes S1 to S3 and SF1) or replicon transcribed from templates that had been subjected to one round (lanes S4 and S7), two rounds (lanes S8 and SF2), three rounds (lanes S5 and S9), or four rounds (S6 and S10) of mutagenesis. In vitro-transcribed genomic RNA from the two parental replicons (lanes SIN and SFV) was used as a marker for the full-length replicon, and poly(A)-selected mRNA from naive BHK cells was used as a mock control (lanes mock). The blot was hybridized with a probe specific for the neo gene. Expected sizes for vector subgenomic RNAs are 1.2 kb for SIN and 1.65 kb for SFV. (B) Complementation analysis. Expression was measured after introduction of nsP-deleted defective β-Gal replicons into SIN- and SFV-derived Neo-resistant BHK pools. Detection of β-Gal expression was performed using a luminescent β-Gal assay kit (Clontech), and the signal was measured in relative light units (RLU). Data are the means of two independent electroporations; the background reading obtained with naive BHK cells was subtracted from all samples. Images were processed with Adobe Photoshop software.
To confirm that Neo resistance was conferred by replicon RNA, naive BHK cells were electroporated with 5 to 10 μg of poly(A)-selected mRNA extracted from either SIN- or SFV- derived Neo-resistant pools or from naive BHK cells as control. Approximately 48 h postelectroporation, the transfected cells were subjected to G418 selection. Transfection with mRNA from both SIN- and SFV-derived pools rapidly generated high numbers of Neo-resistant cells. In contrast, transfection of control mRNA gave no colonies over an extended period of time.
To determine whether vector RNA was actively replicating in Neo-resistant cells, various cell pools were transfected with defective SIN and SFV replicon RNAs that encoded a β-galactosidase (β-Gal) reporter but had the nsP genes deleted. Amplification and subgenomic transcription of the β-Gal mRNA could occur with these defective vectors only if functional nsPs were provided in trans by replicons already present in the drug-resistant pools. The defective β-Gal replicons were transcribed from plasmids pSINBVdlnsP-βgal (derived from pSINBV-βgal [8] by deleting the BspEI fragments) and pSFV3dlnsP-βgal (derived from pSFV3-βgal [15] [GIBCO-BRL] by deleting the PstI fragments). After transfection of the defective replicons, β-Gal expression was observed in all but one pool (Fig. 1B). This result clearly demonstrated that the variant replicons were actively replicating in cells and provided trans complementation. Pool SF1 did not show demonstrable β-Gal expression, indicating a defect reducing either replication of the variant SFV vector or the vector's ability to initiate subgenomic transcription in trans. Since low levels of subgenomic RNA were observed for SF1 in the Northern analysis (Fig. 1A, lane SF1), the lack of β-Gal expression may be a consequence of reduced subgenomic transcription.
Mapping the adaptive genetic determinants of persistence.
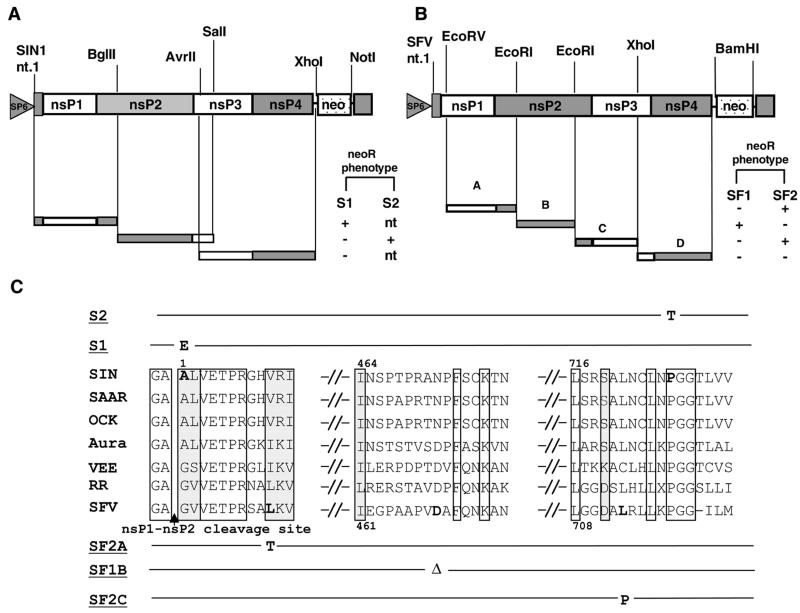
To identify the causal mutations, representative pools S1, S2, SF1, and SF2 were chosen for mapping based on their unique RNA profiles in the Northern analysis. The complete nsP genes of SIN and SFV variant replicons present in these pools were cloned by reverse transcription (RT)-PCR in three and four fragments, respectively (Fig. 2A and B). Each amplified fragment then was substituted for the corresponding fragment in wild-type pSINBV-neo or pSFV-neo, and three independent replicon clones were generated for each nsP fragment substitution. Replicon RNA was transcribed in vitro from the constructs and transfected into naive BHK cells. Following G418 selection, the number of colonies obtained for each construct was compared to the number of colonies obtained with the parental wild-type replicon. For all but one pool, a single specific fragment substitution resulted in the establishment of persistent Neo resistance (Fig. 2A and B). For the SF2 pool, which was derived from vector that had undergone two rounds of mutagenesis, two fragments, SF2A and SF2C, independently conferred the phenotype. Thus, both SIN and SFV replicons that established persistent replication could result from substitution with a defined fragment.
FIG. 2.
Mapping of causal mutations in the variant replicons. (A and B) Schematic illustrations of the cloning strategy used to map the replicon variants. Depicted are the neo-containing SIN and SFV replicons, with corresponding RT-PCR-amplified fragments generated from the Neo-resistant BHK pools. Also shown are nsP coding regions and the restriction sites used in fragment substitutions. nt., nucleotide. The ability of each fragment substitution to efficiently confer Neo resistance (+) compared to the parental vectors (−) is indicated; “nt” denotes fragments not tested. (C) Sequence alignments of the nsP2 regions in which the mutations were located are shown for several alphaviruses. Bold characters indicate amino acid residues where mutations were found; the change is indicated above the alignment for the SIN-derived variants and below the alignment for the SFV-derived variants. In variant SF1B, Δ indicates the deletion of amino acid D469. Since the length of nsP2 varies between SIN and SFV, codon numbering is indicated for each virus. White boxes highlight identical residues among all of the alphaviruses aligned; gray boxes highlight conservative changes. Images were processed with Adobe Photoshop software.
The defined fragments were sequenced entirely and compared to the parental replicon sequence. Each SIN and SFV variant contained only a single amino acid substitution within the nsP2 protein (Fig. 2C). Although the precise location of these amino acid changes differed among the SIN and SFV variants, the amino terminus (amino acid [aa] 1 in variant S1 and aa 10 in variant SF2A) and a small region of the carboxy terminus (aa 726 in variant S2 and aa 713 in variant SF2C) seemed to be targeted preferentially. The latter region is within the putative protease domain of nsP2 (12), where mutation of P726 previously was found to reduce cytopathogenicity of both SIN (7) and a SIN-based replicon (10). Interestingly, the S1 mutation mapped within the nsP1-nsP2 cleavage recognition site (27). Furthermore, the SF1B variant contained an in-frame deletion of aa 469, within a different region of nsP2.
Properties of the cloned variants.
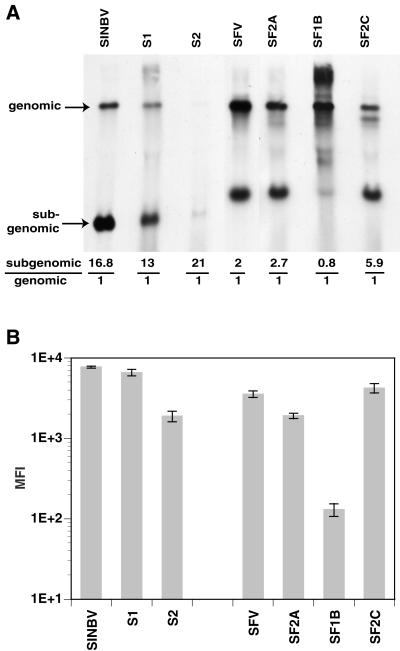
To characterize the cloned replicon variants, we examined the impact of each mutation on RNA replication and heterologous gene expression. The ratios of subgenomic and genomic RNA were evaluated in drug-resistant cell lines obtained using the cloned SIN and SFV replicon variants, as well as naive BHK cells electroporated 2 h earlier with parental replicon RNAs. Cells were labeled with [3H]uridine (100 μCi/ml) in the presence of dactinomycin (1 μg/ml) for 7 h. Total RNA was extracted from the cells and separated by gel electrophoresis (19), the gels were treated and exposed to film (10), and the genomic and subgenomic RNA bands were excised and subjected to scintillation counting (Fig. 3A). Although a direct comparison could not be made with the transiently transfected parental vectors, the individual variant replicons clearly showed differences in molar ratios of subgenomic to genomic RNA among each other. This result suggested that the nsP2 mutations affected the levels of genomic replication and/or subgenomic transcription and that variants S2 and SF2C had smaller amounts of genomic RNA than other variants. A longer exposure confirmed the presence of both genomic and subgenomic RNAs in S2 (data not shown).
FIG. 3.
Analysis of RNA replication and heterologous gene expression. (A) Stable BHK cell lines derived using the cloned variant vectors (lanes S1, S2, SF2A, SF1B, and SF2C) and naive BHK cells electroporated with parental vector RNAs (lanes SINBV and SFV) were labeled with [3H]uridine. Total RNA was extracted and quantitated, and equivalent amounts were run in an agarose-formaldehyde gel. Molar ratios of subgenomic to genomic RNA were measured by scintillation counting of excised gel fragments. Size differences between SIN and SFV vectors are reflected in the subgenomic RNA size. Exposure time for the transfected SINBV and SFV control lanes was shorter than for the drug-resistant cell lanes. (B) BHK cells were electroporated with parental and variant vector RNA encoding E-GFP. At 24 h posttransfection, the cells were analyzed by flow cytometry, and mean fluorescence intensity (MFI) of the GFP-positive cell population was plotted. Data are the means of four independent electroporations, with the standard deviations shown (n = 4). The background MFI reading from mock-transfected BHK cells was subtracted from all the samples. Images were processed with Adobe Photoshop software.
The level of heterologous gene expression among variants and parental replicons was next compared by using replicons expressing the E-GFP (enhanced green fluorescent protein) reporter gene (Clontech). BHK cells were electroporated with in vitro-transcribed replicon RNA, and the level of GFP expression was assayed 24 h later by flow cytometry (Fig. 3B). Although transfection efficiency varied among replicons, GFP expression levels within individual transfected cells were similar to or slightly lower than levels in the parental replicons for most variants. In contrast to all other variants, SFV1B showed inefficient expression of GFP. Since the subgenomic-to-genomic RNA ratio was lower for SF1B than for other variants (Fig. 3A), the observed defect may be a consequence of low subgenomic transcription levels.
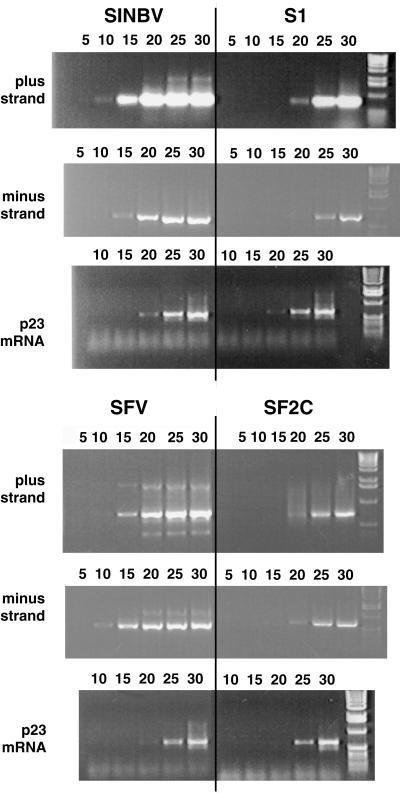
We then analyzed whether the mutations differentially affected plus-strand or minus-strand RNA synthesis. To differentiate the levels of each RNA species, semiquantitative RT-PCR was performed on equivalent amounts of total RNA extracted from either Neo-resistant BHK cell lines containing the cloned SIN and SFV variant replicons or naive BHK cells electroporated 24 h earlier with the parental replicons. Oligonucleotides complementary to either plus- or minus-strand RNA were used for detection of strand-specific cDNA. After cDNA synthesis and RNase A treatment, a 700-bp fragment corresponding to a region of either nsP4 for the SIN variants or nsP3 for the SFV variants was amplified by PCR. Each PCR mixture was divided into multiple aliquots, and one aliquot was analyzed every five amplification cycles (Fig. 4). Both plus- and minus-strand RNA levels were found to be lower with both the S1 and SF2C variants than with the parental vectors at 24 h postelectroporation. Similar results were obtained with the other variants (data not shown). A specific fragment of the housekeeping gene BHKp23 (18) also was synthesized from each sample as an internal standard, and similar amounts of product were obtained in all cases (Fig. 4). This result clearly demonstrated that each variant had ongoing minus-strand synthesis, which is a requirement for persistent replication.
FIG. 4.
Detection of vector minus-strand and plus-strand RNA by RT-PCR. The cDNA corresponding to either minus strand or plus strand was amplified using strand-specific primers from total RNA of either BHK cells lines containing the cloned variants (S1 and SF2C) or naive BHK cells electroporated 24 h earlier with the parental vectors (SINBV and SFV). As an internal control, the housekeeping BHKp23 mRNA was amplified from each sample (p23). Amplification mixtures were divided into six aliquots, and one aliquot per sample was removed after 5, 10, 15, 20, 25, and 30 amplification cycles, as indicated above the lanes. Images were processed with Adobe Photoshop software.
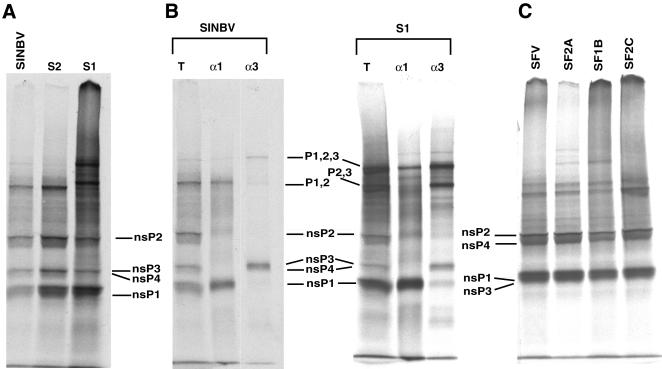
Alphavirus nsPs are translated initially as polyproteins, P1234 and P123 in SIN and P1234 in SFV. These polyproteins are processed subsequently into mature monomers by the nsP2 protease (5, 12), with the processing intermediates playing an important role in the early events of RNA replication, including a shift from minus-strand to plus-strand synthesis (23, 27). Since minus-strand synthesis was maintained with the SIN and SFV variant replicons, we analyzed the effects of the mutations on polyprotein processing. Coupled transcription-translation of parental and variant replicon RNA was performed with rabbit reticulocyte lysates (TnT; Promega) in the presence of [35S]methionine. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis revealed that although all mutants accumulated the nsP monomers, mutants S1, SF2A, and SF1B also accumulated significant amounts of higher-molecular-weight products (Fig. 5A and C). Immunoprecipitation of the in vitro-translated products from SINBV and S1, with antisera specific for either nsP1 or nsP3, indicated that variant S1 accumulated the P123 and P23 precursors (Fig. 5B). In contrast to previous results for mutation of nsP2 residue 726 (10), we did not observe any significant processing difference between variant S2 and its parental replicon. However, several amino acid differences exist among nsPs of the parental replicons used by each laboratory, and these differences in genetic background may affect the processing efficiency. Nonetheless, these results suggested that the maintenance of minus-strand synthesis may be achieved through altered polyprotein processing.
FIG. 5.
In vitro processing of nonstructural polyprotein. (A and C) Coupled transcription-translation of parental and variant replicon RNA was performed in the presence of [35S]methionine, in rabbit reticulocyte lysates, and the products were analyzed by SDS-PAGE on an 8% gel. (B) The translation products (lanes T) of SINBV and variant S1 were immunoprecipitated with polyclonal antibodies against either SIN nsP1 (lanes α1) or SIN nsP3 (lanes α3). Images were processed with Adobe Photoshop software.
Finally, we examined the ability of the variant replicons to be packaged into virion-like particles by supplementing the structural proteins in trans from separate capsid- and glycoprotein-defective helper RNAs transcribed in vitro (16). Interestingly, and in contrast to previous studies, some variant replicons could be packaged as efficiently as the parental replicon (SINBV-GFP [5 × 108 IU/ml] versus S1-GFP [3.8 × 108 IU/ml]) and some with a slightly decreased efficiency (SFV3LacZ [3.8 × 108 IU/ml] versus SF2ALacZ [5 × 107 IU/ml] and SF2CLacZ [107 IU/ml]). The other replicons were packaged at very low efficiency (S2-GFP and SF1BLacZ, ≤le4 IU/ml).
This report extends previous studies on noncytopathic SIN replicon variants (1, 10) with the demonstration that mutations in multiple regions of nsP2 can lead to the establishment of persistent RNA replication. Significantly, similar variants also may be generated with SFV-derived replicons. For both SIN and SFV, regions of nsP2 encompassing the amino terminus or proximal to the carboxy terminus seem to be preferential targets for mutations leading to persistent replication and maintenance of heterologous gene expression.
Alphavirus replicon vectors have many attractive features, and the addition of either long-term expression options or decreased cytopathogenicity should facilitate the expansion of their applications. Maintenance of ongoing minus-strand synthesis and inhibition of cytopathogenicity are requirements for these vectors to establish persistent replication with continued high-level transgene expression. In the alphavirus replication cycle, minus-strand RNA synthesis occurs only during the first few hours postinfection (21, 24). Extensive work (reviewed in references 23 and 27) supports proposed models in which both processing intermediates and mature nsP monomers form alphavirus polymerases with different activities. Final cleavage of the P23 intermediate converts the polymerase activity from synthesis of both minus and plus strands to only plus-strand genomic and subgenomic RNA synthesis (23, 27). Since nsP2 contains the protease domain responsible for the nsP maturation (12), it is noteworthy that all noncytopathic variants characterized to date (this work and reference 10) result from mutation of nsP2. Interestingly, one variant (S1) accumulates the P123 and P23 processing intermediates, indicating that maintenance of minus-strand synthesis may be achieved by deregulating the switch from minus strand to plus strand through this pathway. Similar to previously published studies where temperature-sensitive nsP2 mutants did not abolish or resumed minus-strand synthesis at the nonpermissive temperature (22, 28), one variant in the present study showed accumulation of unprocessed nsP and severe inhibition of subgenomic RNA synthesis. However, most noncytopathic variants (this work and reference 10) displayed high subgenomic RNA expression, indicating that maintenance of minus-strand synthesis through this pathway does not necessarily result in inhibition of subgenomic RNA synthesis. Interestingly, this phenotype is similar to that of another temperature-sensitive SIN mutant, 24R1, in which a mutation in nsP4 permitted continuation of minus-strand synthesis without affecting subgenomic RNA synthesis (20).
The loss of cytopathogenicity was correlated with a reduction of RNA replication in a panel of nsP2 Pro726 mutants (10). Unfortunately, diminished replication also was associated with severely decreased replicon packaging efficiency. One of our variants, S2, which was equivalent to previously isolated variants (10), could not be packaged efficiently into virion-like particles. In contrast, several of our newly identified variants (S1, SF2A, and SF2C) both maintained high-level transgene expression and also were packaged efficiently, thus increasing the versatility of these replicons. For example, the new replicons might be particularly useful for extending expression studies in hippocampal slice cultures (9) without perturbation of host cell metabolism. The degree of cytopathogenicity and the mechanisms by which induction of apoptosis is either inhibited or delayed remain to be established. Importantly, this panel of variants provides a basis for further studies in both cultured cells and animal models of human disease.
REFERENCES
- 1.Agapov E V, Frolov I, Lindenbach B D, Pragai B M, Schlesinger S, Rice C M. Noncytopathic Sindbis virus RNA vectors for heterologous gene expression. Proc Natl Acad Sci USA. 1998;95:12989–12994. doi: 10.1073/pnas.95.22.12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berglund P, Smerdou C, Fleeton M N, Tubulekas I, Liljestrom P. Enhancing immune responses using suicidal DNA vaccines. Nat Biotechnol. 1998;16:562–565. doi: 10.1038/nbt0698-562. [DOI] [PubMed] [Google Scholar]
- 3.Boorsma M, Nieba L, Koller D, Bachmann M F, Bailey J E, Renner W A. A temperature-regulated replicon-based DNA expression system. Nat Biotechnol. 2000;18:429–432. doi: 10.1038/74493. [DOI] [PubMed] [Google Scholar]
- 4.Bredenbeek P J, Frolov I, Rice C M, Schlesinger S. Sindbis virus expression vectors: packaging of RNA replicons by using defective helper RNAs. J Virol. 1993;67:6439–6446. doi: 10.1128/jvi.67.11.6439-6446.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding M X, Schlesinger M J. Evidence that Sindbis virus nsP2 is an autoprotease which processes the virus nonstructural polyprotein. Virology. 1989;171:280–284. doi: 10.1016/0042-6822(89)90539-4. [DOI] [PubMed] [Google Scholar]
- 6.Driver D A, Latham E M, Polo J M, Belli B A, Banks T A, Chada S, Brumm D, Chang S M, Mento S J, Jolly D J, Dubensky T W., Jr Layered amplification of gene expression with a DNA gene delivery system. Ann N Y Acad Sci USA. 1995;772:261–264. doi: 10.1111/j.1749-6632.1995.tb44754.x. [DOI] [PubMed] [Google Scholar]
- 7.Dryga S A, Dryga O A, Schlesinger S. Identification of mutations in a Sindbis virus variant able to establish persistent infection in BHK cells: the importance of mutation in the nsP2 gene. Virology. 1997;228:74–83. doi: 10.1006/viro.1996.8364. [DOI] [PubMed] [Google Scholar]
- 8.Dubensky T W, Jr, Driver D A, Polo J M, Belli B A, Latham E M, Ibanez C E, Chada S, Brumm D, Banks T A, Mento S J, Jolly D J, Chang S M W. Sindbis virus DNA-based expression vectors: utility for in vitro and in vivo gene transfer. J Virol. 1996;70:508–519. doi: 10.1128/jvi.70.1.508-519.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehrengruber M U, Lundstrom K, Schweitzer C, Heuss C, Schlesinger S, Gahwiler B H. Recombinant Semliki Forest virus and Sindbis virus efficiently infect neurons in hippocampal slice cultures. Proc Natl Acad Sci USA. 1999;96:7041–7046. doi: 10.1073/pnas.96.12.7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frolov I, Agapov E, Hoffman J T A, Pragai B M, Lippa M, Schlesinger S, Rice C M. Selection of RNA replicons capable of persistent noncytopathic replication in mammalian cells. J Virol. 1999;73:3854–3865. doi: 10.1128/jvi.73.5.3854-3865.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffin D E, Hardwick J M. Regulators of apoptosis on the road to persistent alphavirus infection. Annu Rev Microbiol. 1997;51:565–592. doi: 10.1146/annurev.micro.51.1.565. [DOI] [PubMed] [Google Scholar]
- 12.Hardy W R, Strauss J H. Processing the nonstructural polyproteins of Sindbis virus: nonstructural proteinase is in the C-terminal half of nsP2 and functions both in cis and in trans. J Virol. 1989;63:4653–4664. doi: 10.1128/jvi.63.11.4653-4664.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herweijer H, Latendresse J, Williams P, Zhang G, Danko I, Schlesinger S, Wolff J. A plasmid-based self-amplifying Sindbis virus vector. Hum Gene Ther. 1995;6:1161–1167. doi: 10.1089/hum.1995.6.9-1161. [DOI] [PubMed] [Google Scholar]
- 14.Levine B, Goldman J E, Jiang H H, Griffin D E, Hardwick J M. Bcl-2 protects mice against fatal alphavirus encephalitis. Proc Natl Acad Sci USA. 1996;93:4810–4815. doi: 10.1073/pnas.93.10.4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liljeström P, Garoff H. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. BioTechnology. 1991;9:1356–1361. doi: 10.1038/nbt1291-1356. [DOI] [PubMed] [Google Scholar]
- 16.Polo J M, Belli B A, Driver D A, Frolov I, Sherrill S, Hariharan M J, Townsend K, Perri S, Mento S J, Jolly D J, Chang S M, Schlesinger S, Dubensky T W., Jr Stable alphavirus packaging cell lines for Sindbis virus and Semliki Forest virus-derived vectors. Proc Natl Acad Sci USA. 1999;96:4598–4603. doi: 10.1073/pnas.96.8.4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pushko H, Parker M, Ludwig G V, Davis N L, Johnston R E, Smith J F. Replicon-helper systems form attenuated Venezuelan equine encephalitis virus: expression of heterologous genes in vitro and immunization against heterologous pathogens in vivo. Virology. 1997;239:389–401. doi: 10.1006/viro.1997.8878. [DOI] [PubMed] [Google Scholar]
- 18.Rojo M, Pepperkok R, Emery G, Kellner R, Stang E, Parton R G, Gruenberg J. Involvement of the transmembrane protein p23 in biosynthetic protein transport. J Cell Biol. 1997;139:1119–1135. doi: 10.1083/jcb.139.5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 20.Sawicki D, Barkhimer D B, Sawicki S G, Rice C M, Schlesinger S. Temperature sensitive shut-off of alphavirus minus strand RNA synthesis maps to a nonstructural protein, nsP4. Virology. 1990;174:43–52. doi: 10.1016/0042-6822(90)90052-s. [DOI] [PubMed] [Google Scholar]
- 21.Sawicki D L, Sawicki S G. Short-lived minus-strand polymerase for Semliki Forest virus. J Virol. 1980;34:108–118. doi: 10.1128/jvi.34.1.108-118.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sawicki D L, Sawicki S G. A second nonstructural protein functions in the regulation of alphavirus negative-strand RNA synthesis. J Virol. 1993;67:3605–3610. doi: 10.1128/jvi.67.6.3605-3610.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sawicki D L, Sawicki S G. Role of the non-structural polyproteins in alphaviral RNA synthesis. Adv Exp Med Biol. 1998;440:187–198. doi: 10.1007/978-1-4615-5331-1_23. [DOI] [PubMed] [Google Scholar]
- 24.Sawicki D L, Sawicki S G, Keranen S, Kaariainen L. Specific Sindbis virus-coded function for minus-strand RNA synthesis. J Virol. 1981;39:348–358. doi: 10.1128/jvi.39.2.348-358.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlesinger S, Dubensky T M., Jr Alphavirus vectors for gene expression and vaccines. Curr Opin Biotechnol. 1999;10:434–439. doi: 10.1016/s0958-1669(99)00006-3. [DOI] [PubMed] [Google Scholar]
- 26.Stollar V. Togaviruses in cultured arthropod cells. In: Schlesinger R W, editor. The togaviruses—biology, structure, replication. New York, N.Y: Academic Press, Inc.; 1980. pp. 584–621. [Google Scholar]
- 27.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suopanki J, Sawicki D L, Sawicki S G, Kaariainen L. Regulation of alphavirus 26S mRNA transcription by replicase component nsP2. J Gen Virol. 1998;79:309–319. doi: 10.1099/0022-1317-79-2-309. [DOI] [PubMed] [Google Scholar]
- 29.Weiss B, Rosenthal R, Schlesinger S. Establishment and maintenance of persistent infection by Sindbis virus in BHK cells. J Virol. 1980;33:463–474. doi: 10.1128/jvi.33.1.463-474.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiong C, Levis R, Shen P, Schlesinger S, Rice C M, Huang H V. Sindbis virus: an efficient, broad host range vector for gene expression in animal cells. Science. 1989;243:1188–1191. doi: 10.1126/science.2922607. [DOI] [PubMed] [Google Scholar]