Figure 3. Time-course Transcriptomic Analysis of hPSC Differentiation into Ventral Diencephalon and Hypothalamic Arcuate Neurons.
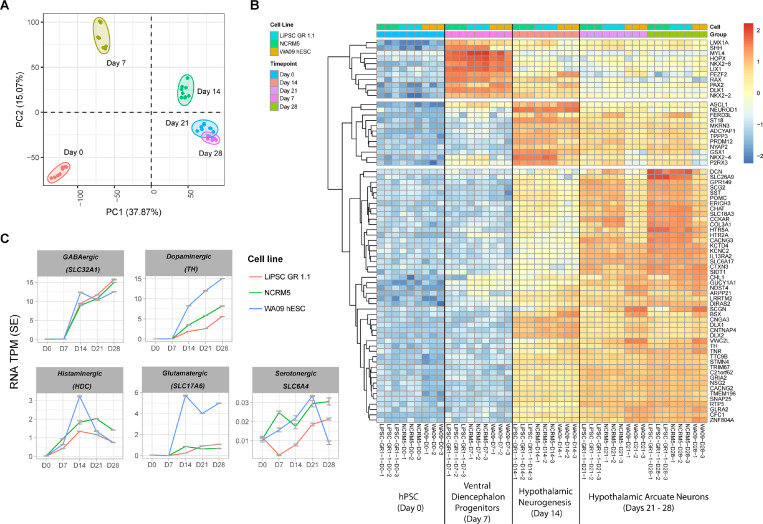
(A) PCA of a time-course mRNA-seq experiment including samples from three independent cell lines. Note the clustering of the samples according to the time-point of differentiation, suggesting the same developmental trajectory between samples from different cell lines.
(B) The heatmap represents the top 20 gene markers upregulated (log2FC) at days 7, 14, and 21, and the top 60 gene markers upregulated at day 28, relative to DIV 0 (hPSC). Due to the overlap of top markers between the time points, the total number of genes represented on the heatmap is 71. Note the stepwise upregulation of ventral diencephalon markers RAX, NKX2.1, ASCL1, DLX1, and ARX, as well as arcuate nucleus markers ISL1, POMC, SST, and TH. N = 3 independent hPSC lines differentiation experiments for all datasets presented (LiPSC-GR 1.1, WA09, NCRM5).