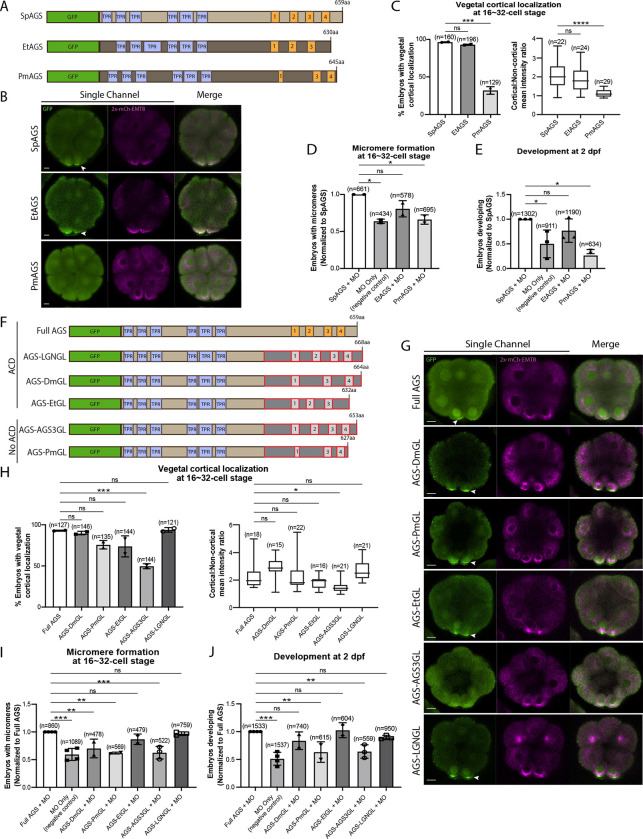
Figure 5. Molecular evolution of AGS C-terminus facilitates micromere formation.
A, Design of GFP-AGS constructs from three different species tested in this study, namely, S. purpuratus (Sp), E. tribuloides (Et), and P. miniata (Pm). TPR motifs are marked in blue, and GL motifs are in orange. B¸ Single Z-slice confocal images of sea urchin embryos at 16-cell stage showing localization of each GFP-AGS. Embryos were injected with 0.3μg/μl stock of GFP-AGS mRNAs and 0.25μg/μl stock of 2x-mCherry-EMTB mRNA. The white arrowhead indicates vegetal cortical localization of GFP-AGS. C, % of embryos with vegetal cortical localization of GFP-AGS (left) and the ratio of the cortical-to-non-cortical mean intensity (right) in 16~32-cell embryos. Statistical analysis was performed against SpAGS by One-Way ANOVA. D-E, Embryos were injected with 0.15μg/μl stock of GFP-AGS mRNAs and 0.75mM SpAGS MO. The number of embryos making micromeres (D) and developing to pluteus stage (E) were scored and normalized to that of the Full AGS. Statistical analysis was performed by One-Way ANOVA. F, Design of GFP-AGS C-terminal chimeric mutant constructs tested in this study. TPR motifs are marked in blue, and GL motifs are in orange. The brown section shows the SpAGS portion, and the red and dark grey boxes show the non-sea urchin (non-SpAGS) C-terminal sequence introduced. Protein sequences used include Drosophila Pins (Dm), P. miniata AGS (Pm), E. tribuloides AGS (Et), H. sapiens AGS3 (AGS3) and H. sapiens LGN (LGN). G¸ Single Z-slice confocal images of sea urchin embryos at 8~16-cell stage showing localization of each GFP-AGS. Embryos were injected with 0.3μg/μl stock of GFP-AGS mRNA and 0.25μg/μl stock of 2x-mCherry-EMTB mRNA. The white arrowheads indicate vegetal cortical localization of GFP-AGS. H, % of the embryos with vegetal cortical localization of GFP-AGS chimeric mutants (left) and the ratio of the cortical-to-non-cortical mean intensity (right) in 16~32-cell embryos. Statistical analysis was performed against Full AGS by One-Way ANOVA. I-J, Embryos were injected with 0.15μg/μl stock of GFP-AGS mRNAs and 0.75mM SpAGS MO. The number of embryos making micromeres (I) and developing to gastrula or pluteus stage (J) were scored and normalized to that of the Full AGS. Statistical analysis was performed by One-Way ANOVA. n indicates the total number of embryos scored. * represents p-value < 0.05, ** p-value < 0.01, *** p-value < 0.001, and **** p-value <0.0001. Each experiment was performed at least two independent times. Error bars represent standard error. Scale bars=10μm.