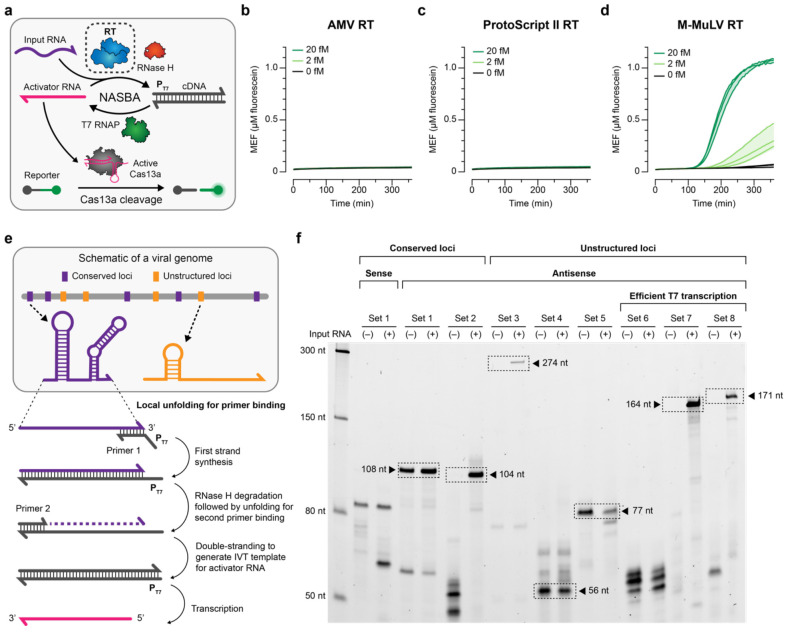
Fig. 1 ∣. In-house NASBA formulation provides flexibility for reaction optimization.
a, Schematic overview of NASBA, which uses cycles of reverse transcription, RNase H-mediated degradation and T7 transcription to convert and amplify an input RNA into an antisense activator RNA. The antisense activator serves as an input to Cas13a-based detection which generates a fluorescent output signal. In-house NASBA formulation enables screening of different reverse transcriptases (RTs). One-pot in-house NASBA-Cas13a targeting the ORF1ab of the SARS-CoV-2 genome, with 0, 2 or 20 fM synthetic SARS-CoV-2 genome and 1 U/μL of b, avian myeloblastosis virus (AMV) RT, c, ProtoScript II RT, or d, Moloney murine leukemia virus (M-MuLV) RT. Readout was observed only with M-MuLV RT. e, Schematic of the steps in NASBA with a cartoon of viral genome structures that could influence where NASBA primers bind and impact NASBA efficiency. f, To test different primer sets, RNA products were extracted from one- pot NASBA (lacking Cas13a) and analyzed by urea-PAGE. Reactions were initiated using 2.5 U/μL M-MuLV RT with 0 (−) or 20 (+) fM synthetic SARS-CoV-2 genome. The expected RNA product for each primer set is boxed and its length is indicated, unless the band was not present as in the case of sets 1 and 6 (expected products 104 nt and 164 nt, respectively). Data in b–d are n=3 independent biological replicates, each plotted as a line with raw fluorescence standardized to MEF. Shading in b–d indicates the average of the replicates ± standard deviation. Data in f are one representative of n=3 independent biological replicates; the other replicates and the uncropped, unprocessed image in f are in Supplementary Data 2. Sequences of primers and gRNAs are listed in Supplementary Data 1.