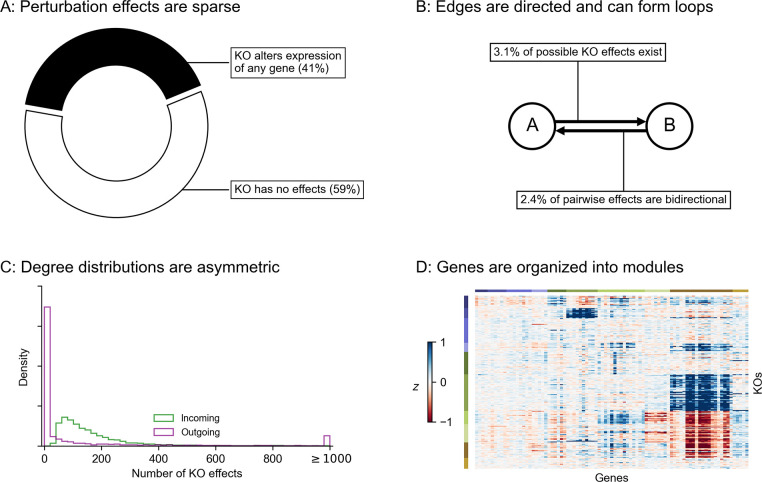
Figure 1: Key properties of gene regulatory networks.
Data from Replogle et. al., 2022. (A) Of the 5,247 perturbations in our analysis subset, 2,152 (41%) have a measurable effect on the transcriptional state of cells (energy-test p < 0.001). (B) Among all ordered pairs of genes, 3.1% (865,719 pairs) have a one-directional effect (FDR-corrected p < 0.05). Of these pairs with KO effects, 2.4% (20,621 pairs) further have bidirectional effects. (C) Summaries of the distribution of KO effects (Anderson-Darling p < 0.05) from the perspective of genes as subject to perturbation (outgoing effects) and as target genes when other genes are perturbed (incoming effects). (D) Subset of z-normalized expression data corresponding to 10 gene modules, using labels as provided in the dataset – each modules is labeled by a color in the to bars above the x- and y-axes, and z-scores are clipped at ±1, for visualization.