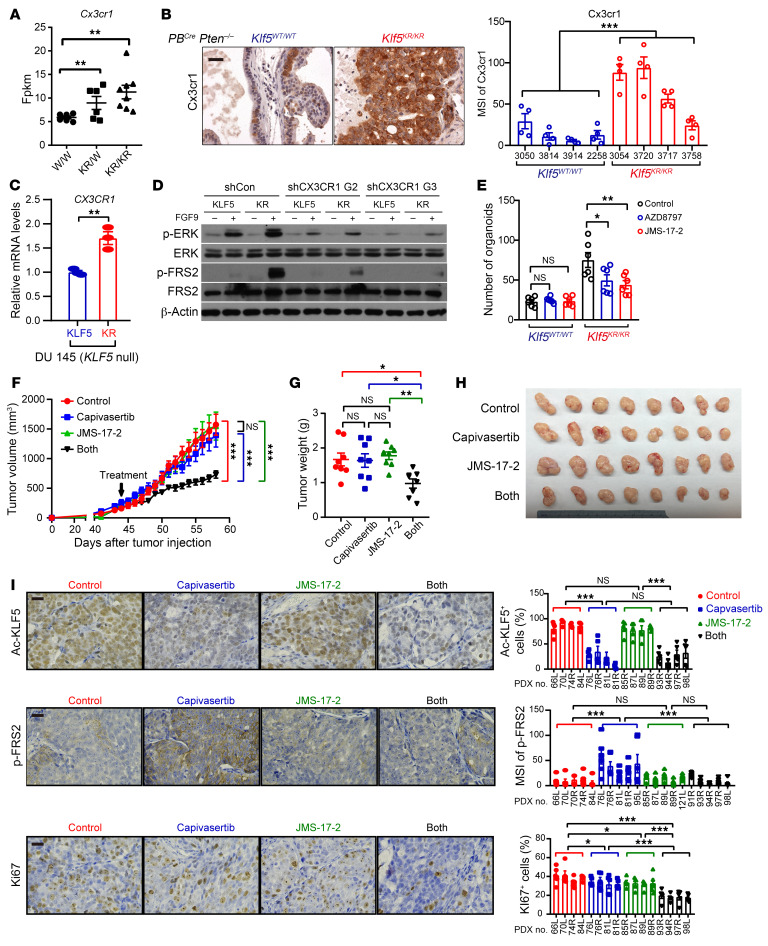
Figure 8. Klf5 deacetylation upregulates CX3CR1 to enhance FGFR1 signaling activity, and blocking CX3CR1 sensitizes tumor cells to AKT inhibition.
(A and B) The expression levels of Cx3cr1 were higher in PBCre Pten–/– Klf5KR/KR prostate tumors, as indicated by RNA-Seq (A) and IHC staining (B). W/W, PBCre Pten–/– Klf5WT/WT; KR/W, PBCre Pten–/– Klf5WT/KR; KR/KR, PBCre Pten–/– Klf5KR/KR. Scale bar: 50 μm. (C) Expression of CX3CR1 mRNA in DU 145 prostate cancer cells with KLF5WT (KLF5) or KLF5KR (KR) by real-time qPCR. (D) DU 145 cells expressing KLF5WT (KLF5) and KLF5KR (KR) were treated with FGF9 (50 ng/mL) for 5 minutes. FGFR1 downstream p-ERKThr202/Tyr204 and p-FRS2Tyr436 were detected by Western blotting. G2 and G3 are 2 shRNAs of CX3CR1. shCon, control shRNA. (E) Inhibitors of CX3CR1 selectively suppressed the organoid formation of mouse prostate cancer cells with the Klf5KR mutant in the context of Pten deficiency. AZD8797 (50 nM) and JMS-17-2 (1 nM) are 2 different CX3XR1 inhibitors. (F–H) PTEN-deficient PDXs (The Jackson Laboratory, TM00298) on NSG mice were treated daily with the AKT inhibitor capivasertib and/or the CX3CR1 inhibitor JMS-17-2 as indicated. JMS-17-2 sensitized the effects of capivasertib on PDX growth, as indicated by the tumor volumes at different time points (F), tumor weights (G), and images (H) at excision. (I) The expression levels of Ac-KLF5, p-FRS2, and Ki67 were evaluated by IHC staining and quantitative analysis. Scale bars: 50 µm. *P < 0.05, **P < 0.01, and ***P < 0.001, by 2-tailed Student’s t test (A, C, E, and G) and 2-way ANOVA (B, F and I).