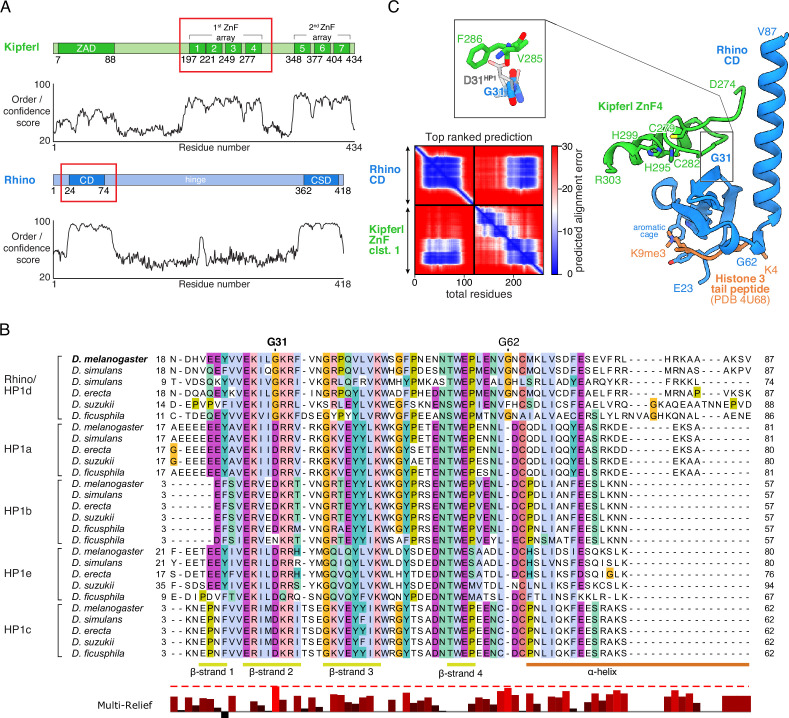
Figure 1. Structure prediction and phylogenetic analyses point to a Rhino-specific residue involved in binding Kipferl.
(A) Domain organization of Kipferl and Rhino, with the AlphaFold2 Multimer predicted local distance difference test (pLDDT) score plotted as a measure of order or disorder alongside. Red boxes indicate the smallest interacting fragments identified by yeast two-hybrid experiments by Baumgartner et al., 2022. ZAD, zinc finger-associated domain; ZnF, zinc finger; CD, chromodomain; CSD, chromo shadow domain. (B) Multiple sequence alignment of heterochromatin protein 1 (HP1) family proteins in five selected species harboring an unequivocally identified Kipferl homolog (see Figure 1—figure supplement 2). Rhino-specific amino acid residues are indicated. Protein accessions and identifiers are documented in Supplementary file 1. Multi-Relief representation indicates residues that differ significantly in Rhino homologs versus other HP1 variant proteins. Note that two Rhino paralogs are identified in D. simulans (see Supplementary file 1 for accessions). (C) Predicted aligned error (PAE) plot for the top ranked AlphaFold2 Multimer prediction of the Rhino chromodomain with the Kipferl ZnF cluster 1 (left) and structure of the complex in cartoon representation (Rhino in blue; Kipferl in green), together with the H3K9me3 peptide (orange) as observed in a Rhino–H3K9me3 crystal structure (PDB ID 4U68). Key residues of Rhino’s aromatic cage and H3K9me3, as well as of Kipferl’s C2H2 ZnF4 are shown in sticks representation. Only the interacting ZnF4 is shown. Depicted in the inset are Rhino G31 and HP1 D31, with HP1 (PDB ID 6MHA) superimposed on Rhino chromodomain residues 26–57 (root mean square deviation = 0.55 Å), together with Kipferl V285 and F286, illustrating that D31 would lead to steric clashes with Kipferl.