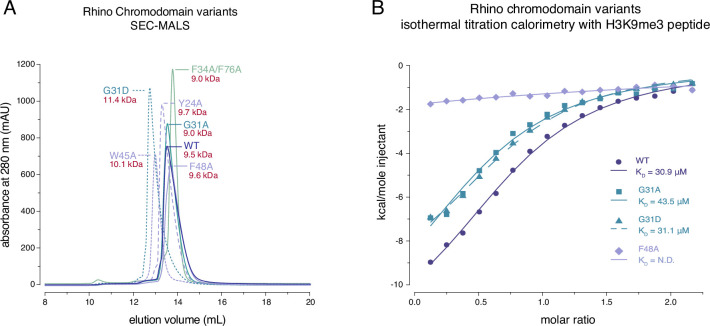
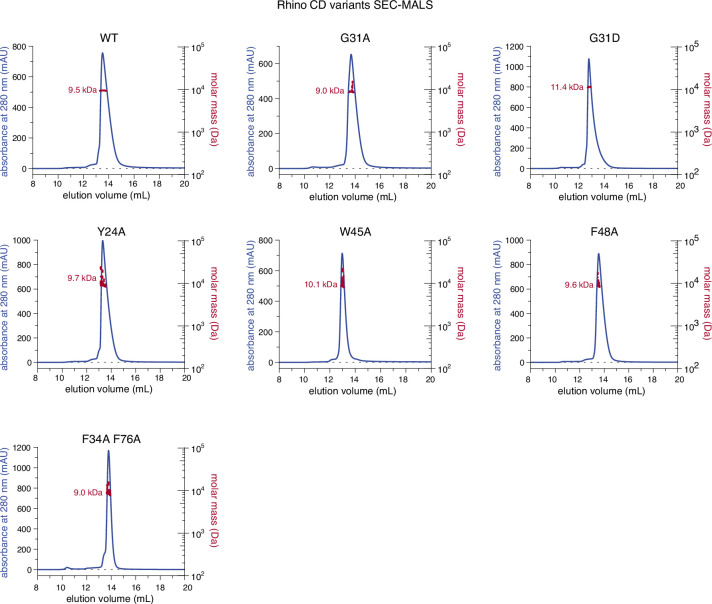
Figure 2. Rhino G31 point mutations do not affect Rhino’s ability to bind H3K9me3.
(A) Line graph summarizing size-exclusion chromatography with inline multiangle light scattering (SEC-MALS) results for the examined Rhino chromodomain constructs. The in solution molecular weight is indicated for each construct. (B) Isothermal titration calorimetry results showing the binding of indicated Rhino chromodomain constructs to the H3K9me3-modified histone tail peptide.