Dear Editor,
The tumor suppressor TP53 is the most frequently mutated gene in cancer [1]. Across diverse myeloid malignancies TP53 disruption is common and associated with poor-risk disease [2] and therapeutic resistance [3]. Unusually, TP53 mutations are rare in chronic myelomonocytic leukemia (CMML) [4, 5], despite disease features otherwise overlapping with myelodysplastic syndromes (MDS), myeloproliferative neoplasms and acute myeloid leukemia (AML): each of which harbors sizeable proportions of TP53-mutated cases. Recently, the largest cohort to date reported TP53 mutations in only 2.4% of 1315 CMML cases [4]. Predictably these patients displayed adverse features and inferior AML-free survival (LFS) and overall survival (OS) compared with TP53 wild-type (TP53WT) CMMLs.
While the clinical characteristics of TP53-mutated CMML are now comprehensively described [4], this represents a small fraction of CMML cases. Additionally, p53 activity can be modulated by non-mutational mechanisms, for example via altered transcriptional expression, posttranslational modifications, and cellular localization [6]. We hypothesized that p53 dysfunction might otherwise characterize a hitherto-unknown subset of TP53WT CMML patients, and so investigated TP53 mutations, allelic status, expression level, and therapeutic response in a large international collaborative CMML cohort.
We studied 648 CMML patients from North−West England with available clinical, mutational, and outcome data. Subsets of patients treated at The Christie (Manchester, UK) underwent RNA-sequencing on bone marrow (BM) CD34-sorted hematopoietic stem/progenitor cells (HSPCs; n = 33); and p53 immunohistochemical (IHC) staining on archived BM trephine samples (n = 31; n = 14 overlapping both cohorts). Separately, we analyzed 92 patients treated at National Taiwan University Hospital (NTUH, Taipei, Taiwan) for whom presentation BM mononuclear cells (MNCs; n = 92) and RNA-sequencing (n = 90/92) data were available [7]. Finally, we re-analyzed published RNA-sequencing data from BM MNCs of 24 patients from Hospital Morales Meseguer (Murcia, Spain) [8]. Each cohort included healthy BM controls (HCs). Additional cohort details and experimental methods are provided as Supplementary Data (Methods S1−8, Tables S1−3).
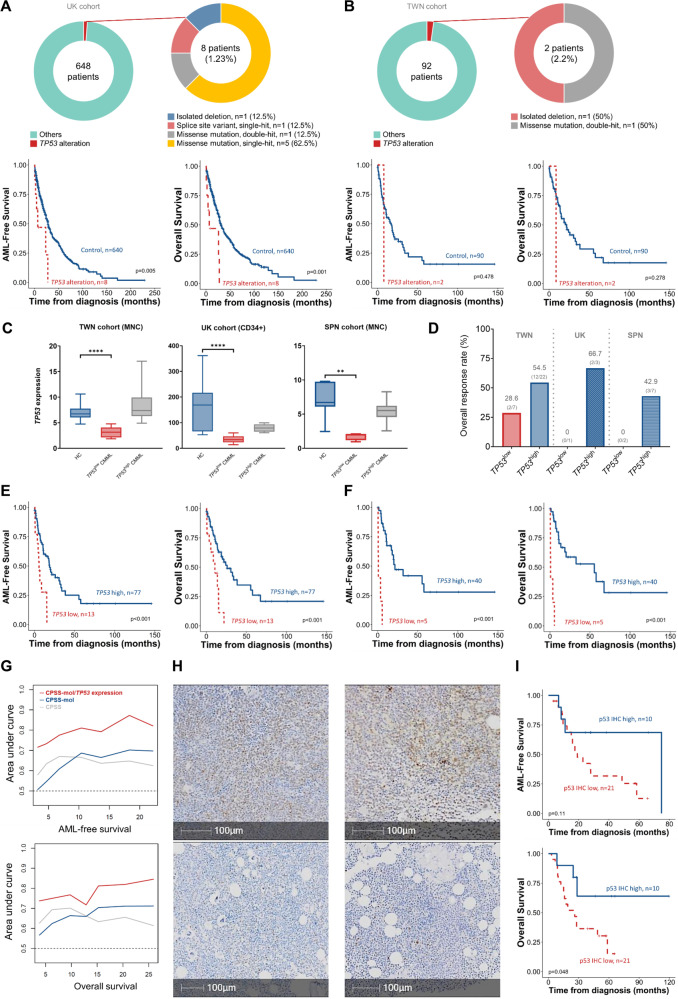
Median ages of the 648 UK and 92 Taiwanese patients were 75 and 71 years, respectively, both with male predominance (Tables S4−5). Only eight (1.23%) and two (2.2%), respectively, carried TP53 alterations (Fig. 1A, B). As expected, UK TP53-altered patients displayed significantly inferior outcomes compared with TP53WT (Fig. 1A), with a similar trend in the Taiwan cohort (Fig. 1B). Thus, we corroborate the paucity of TP53 mutations in CMML [4] across previously unreported UK and Taiwan cohorts.
Fig. 1. Incidence and prognostic impact of TP53 alterations and the prognostic implication of TP53 expression in CMML.
A Incidence of TP53 alterations in the UK CMML cohort (upper) and the outcomes of patients with or without any type of TP53 alteration (lower). B Incidence of TP53 alterations in the Taiwanese CMML cohort (upper) and the outcomes of patients with or without any type of TP53 alteration (lower). C Box and whisker plots displaying TP53 expression of healthy controls (HC) and CMML patients without TP53 alterations across three cohorts. MNC: mononuclear cells. ****P ≤ 0.0001, **P ≤ 0.01. P values were computed using the Mann–Whitney test. Segregation of patients into TP53low and TP53high subgroups was performed by the maximally selected rank method. D Bar plots showing overall response rates to hypomethylating agent monotherapy in patients with lower and higher TP53 expression. Numbers in brackets denote responders over the total number of individuals in each group. E Low TP53 RNA expression conferred significantly worse acute myeloid leukemia-free survival (LFS) and overall survival (OS) in CMML patients in the Taiwan RNA-sequencing discovery cohort. F TP53 expression significantly discriminated patients’ LFS and OS in the ASXL1 wild-type population in the discovery cohort. G Time-dependent ROC curve analyses demonstrate that TP53 expression can refine and improve current prognostic systems. H Representative bone marrow sections stained by immunohistochemistry (IHC) for p53 expression from CMML patients in the UK cohort. Nuclei with clear brown color regardless of staining intensity were regarded as p53 positive. Two exemplar high (upper row) and two low (lower row) expressors are shown. I Patients with lower p53 IHC expression displayed inferior survival compared to those with higher expression. The cutoff for p53 protein expression (25.4%) distinguishing lower and higher p53 groups was determined using maximally selected rank statistics.
Examining TP53 gene expression levels across different cell types in normal and malignant hematopoiesis (from publicly available datasets) revealed that healthy HSCs express significantly higher TP53 than in MDS (p < 0.01, Fig. S1). Therefore, we explored TP53 expression levels and their clinical significance in TP53WT CMML cases, similarly observing lower TP53 in the UK and Spain cohorts (Fig. S2A). Notably, TP53 expression in HSPCs was significantly higher than in MNCs from both healthy and disease contexts (Fig. S2B).
Our Taiwan RNA-sequencing CMML discovery cohort was stratified into TP53high and TP53low transcriptional expression groups. TP53low patients displayed significantly lower expression than HCs, whereas TP53high expression levels were comparable to controls (Fig. 1C). Thus, while most CMML patients are TP53WT, BM cells from a subset (~15%) display abnormally low TP53 expression, suggesting potential for altered p53 (and downstream) function in these patients. Considering the functional crosstalk between the MDM2-MDMX complex and p53 we examined correlations between MDMX, MDM2, and TP53. TP53low expressors exhibited higher MDMX than TP53high, with no difference observed for MDM2 (Fig. S3). Clinical and mutational features did not differ between the two TP53 expression subgroups (Tables S6−11).
Given the association between TP53 mutations and HMA resistance [3], we examined whether TP53 expression correlated with HMA response in the TP53WT context. Despite limited sample sizes (Table S12), TP53low patients showed a consistent trend towards poorer HMA response rates across cohorts (Fig. 1D). TP53low patients displayed significantly shorter LFS and OS than TP53high cases (Fig. 1E). In subgroup analyses, lower TP53 retained strong predictive value for LFS and OS even within CPSS and CPSS-Molecular-stratified subgroups (Figs. S4, 5), and in exclusively ASXL1WT patients (Fig. 1F). Time-dependent ROC curve analysis revealed potential for TP53 expression to enhance current prognostication systems (Fig. 1G). In multivariable analysis, lower TP53 expression remained prognostically detrimental for LFS and OS (Table S13). This was consistent across validation cohorts (Figs. S6, 7; Tables S14, 15), strengthening the observed link between lower TP53 expression and adverse outcomes.
We subsequently explored TP53 expression at the protein level by IHC in 31 CMML trephine samples (Fig. 1H). We found no significant correlation between p53 IHC and TP53 RNA-sequencing expression levels: albeit with only a small overlapping cohort with both available (Table S16, 17, Fig. S8), and comparing different populations (whole BM vs CD34 +, respectively). In the full IHC cohort, however, low p53 expressing cases exhibited significantly inferior OS (p = 0.048; Fig. 1I), validating our observations comparing TP53 transcript levels.
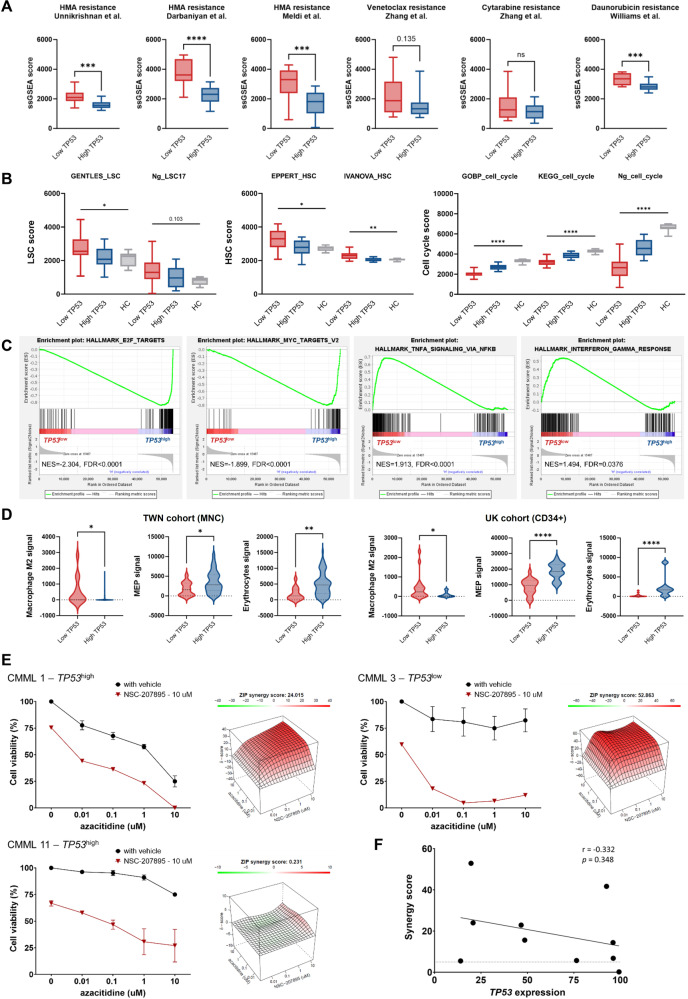
We next sought how lower TP53 expression might influence CMML biology and prognosis. Concordant with our clinical observation (Fig. 1D), single-sample GSEA showed enrichment of HMA resistance signatures in TP53low patients, consistently across cohorts (Figs. 2A, S9). We hypothesized that altered TP53 expression might be associated with aberrant self-renewal and cell cycle programs: recognized mediators of established HMA resistance mechanisms [9]. TP53low cells showed enrichment for LSC and HSC genes, and relative decrease in cell cycle-related genes, as compared with TP53high and HC (Figs. 2B, S10). Thus, TP53low CMML displays distinct stemness and quiescence signatures, linked to poor HMA response in these patients.
Fig. 2. Biological and therapeutic implications of TP53 expression in CMML.
A Box plots displaying resistance signatures derived from single-sample GSEA for hypomethylating agents (HMA), venetoclax, cytarabine, and daunorubicin in patients with lower and higher TP53 expression in the UK CD34 + -sorted cohort. B Box plots displaying scores of leukemic stem cell (LSC), hematopoietic stem cell (HSC), and cell cycle of patients with TP53high and TP53low expression and healthy controls (HC) in the UK CD34 + -sorted cohort. C Representative GSEA plots of pathway enrichment in CMML patients with the lowest 25% vs highest 25% TP53 expression in the UK CD34 + -sorted cohort. D Violin plots displaying different signatures seen in the Taiwan discovery cohort and the UK validation cohort. MEP: megakaryocytic-erythroid progenitors. A, B, D ****P ≤ 0.0001, ***P ≤ 0.001, **P ≤ 0.01, *P ≤ 0.05. P values were computed using Mann–Whitney or Kruskal–Wallis test. E Representative 3D synergy plots using zero interaction (ZIP) model (right) and dose response curves (left) for CMML bone marrow mononuclear cells (n = 3 patients; mean + SEM) treated for 72 h ex vivo with NSC-207895 and azacitidine combination at various concentrations. The presence of synergy was determined utilizing the SynergyFinder computational package and the ZIP synergy index where red denotes synergism and green denotes antagonism. A positive synergy score is the percent more cell death than expected. F Dot plot displaying the correlation between TP53 expression and synergy score of 10 patient samples.
By GSEA TP53low cells exhibited depleted expression of p53-dependent pathways, including MYC targets, G2/M checkpoints, and DNA repair (Figs. 2C, S11). Interestingly, these were all among the most upregulated pathways in TP53-mutant (vs TP53WT) samples across multiple cancers in TCGA data [1], implying that the driving biology of TP53low CMML is distinct from (and not functionally equivalent to) that of oncogenic TP53 mutations. Conversely, TP53low patients demonstrated enhanced TNF-alpha and inflammatory signals (Figs. 2C, S11), highlighting possible crosstalk between p53 and extrinsic factors in the CMML BM microenvironment. Similar results were observed in our other cohorts (Fig. S11). Taken together, compared with HCs or TP53high CMML, TP53low CMML cells display relatively quiescent cell cycle but heightened inflammation.
With emerging evidence suggesting discrete roles for p53 in regulating inflammation and immune cell landscape [10], we applied xCell to our transcriptomic datasets to analyze signals of 22 cell types. TP53low CMML displayed significantly stronger M2-macrophage, but lower megakaryocytic-erythroid progenitors (MEP) signals compared with TP53high (Fig. 2D, Table S18). Interestingly, these findings were consistent in the UK CD34+ dataset, suggesting lineage priming at the progenitor level.
Finally, we explored whether reduced TP53 expression in this CMML subset could be exploited therapeutically. HMAs are the only approved disease-modifying drugs for CMML but often yield disappointing responses [11]. Despite extensive efforts, no combination has yet reported survival advantage over HMA monotherapy. Since the MDM2/MDMX complex degrades wild-type p53, dual inhibition may offer more comprehensive modulation, as suggested by early clinical results in TP53WT AML/MDS following HMA failure [12]. Combining NSC-207895, a dual MDMX/MDM2 inhibitor, and p53 activator, with azacitidine at various concentrations, we observed clear and substantial synergy in primary samples ex vivo from 10/11 patients (Fig. 2E; Table S19). There was a trend towards inverse correlation between TP53 expression and empirical synergy scores (Figs. 2F, S12), suggesting potential for pharmacological p53 activation to enhance HMA sensitivity in CMML with broad efficacy; perhaps preferentially in adverse TP53low expressing cases (although we could not validate this experimentally, lacking availability of matched post-treatment samples).
An intriguing question remains: why are TP53 mutations so infrequent in CMML? Speculatively, TP53 mutations might induce unknown synthetic lethalities in CMML cells; or they may promote alternative lineage specification pathways, re-directing the expressed phenotype and resultant disease classification. Supporting the latter, most studied TP53MUT hematopoietic models report enhanced stemness or propagation of megakaryocytic/erythroid lineage [13], rather than the myelomonocytic expansions that define the CMML phenotype. For example, TP53 knockout synergized with NRASG12D to specifically transform MEPs, but not other HSPC types, in an AML murine model [14]. Accordingly, we observed significant under-representation of myelomonocytic/blastic M4/M5 FAB subtypes associated with TP53 mutations amongst 1511 AML cases at NTUH, and re-analyzing 577 cases from TCGA and BeatAML datasets (odds ratio 0.48 and 0.49, respectively; Table S20). Thus, acquisition of TP53 mutations onto the canonical CMML mutation background might alter the resultant phenotype away from clinicopathological features compatible with CMML diagnostic criteria.
When present, TP53 mutations confer adverse prognosis in CMML as in other cancers. However, our study identifies prognostic implications of mutation-independent TP53 dysregulation in CMML relevant to a much larger minority of patients (~15%). Prior TCGA analysis revealed substantial variation in TP53 expression in both TP53MUT and TP53WT tumors [15], with TP53WT expression lower than in missense but higher than in truncating mutations. Furthermore, the relationship between expression and prognosis differed across cancers [15]. Our data suggest that in CMML TP53 expression level plays a role in dictating disease aggressiveness and therapeutic response, of relatively greater importance than TP53 mutation status in this disease.
In conclusion, ours is the first study to link low TP53 expression with distinct features and outcomes in CMML. We confirm the rarity of TP53 mutations, whilst identifying a novel subgroup with aberrantly low TP53 expression, associated with higher HMA resistance, distinctive biology, and inferior prognosis. We highlight potential for combining HMA and MDMX/MDM2 inhibition to restore HMA sensitivity, as an attractive candidate therapeutic approach for clinical study to address this unmet clinical need.
Supplementary information
Acknowledgements
We acknowledge the service provided by Department of Laboratory Medicine, Department of Medical Research, and Division of Hematology, Department of Internal Medicine, National Taiwan University Hospital and Advanced Imaging and Flow Cytometry Facility, CRUK Manchester Institute, University of Manchester.
Author contributions
YHW was responsible for data collection and management, statistical analysis, interpretation, visualization, literature research, and manuscript writing; KG assisted in bioinformatic and statistical analysis; CCL, CYY, AJ, HAH, and WCC were responsible for data collection, management, and interpretation; and HFT, KB, and DHW conceived and coordinated the study and revised the manuscript.
Funding
The University of Manchester’s Epigenetics of Haematopoiesis Laboratory is core funded by grants from The Oglesby Charitable Trust. DHW is also supported by a Cancer Research UK Advanced Clinician Scientist Fellowship (RCCASF-Nov22/100001) and the University of Manchester Sybil Mary Pilkington Leukaemia Research Fellowship. The work was also supported by grants from Ministry of Science and Technology, Taiwan, project number: MOST 109-2314-B-002-221, 109-2314-B-002-222, 111-2314-B-002-280; and Taiwan Ministry of Health and Welfare, project number: MOHW109-TDU-B-211-134009.
Data sharing and declaration
The data reported in this article will be provided to collaborating investigators through reasonable request to the corresponding authors after requisite institutional review board approval. The data are not publicly available due to privacy or ethical restrictions.
Competing interests
The authors declare no competing interests.
Ethics approval
Use of primary human tissue was in compliance with the ethical and legal framework of the UK’s Human Tissue Act, 2004. Primary samples were from Manchester Cancer Research Centre’s Tissue Biobank instituted with the approval of the South Manchester Research Ethics Committee (18/NW/0092) and licensed by the Human Tissue Authority (license number 30004). Use was authorized following ethical review by the Tissue Biobank’s scientific sub-committee (approval 17_KIBA_01). The NTUH Research Ethics Committee approved the study (#201709072RINC). The study was approved by the institutional review boards of each participating hospital, with informed consent obtained in accordance with the Declaration of Helsinki.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kiran Batta, Email: kiran.batta@manchester.ac.uk.
Daniel H. Wiseman, Email: daniel.wiseman@manchester.ac.uk
Supplementary information
The online version contains supplementary material available at 10.1038/s41408-024-01087-7.
References
- 1.Donehower LA, Soussi T, Korkut A, Liu Y, Schultz A, Cardenas M, et al. Integrated analysis of TP53 gene and pathway alterations in the cancer genome Atlas. Cell Rep. 2019;28:1370–84.e5. doi: 10.1016/j.celrep.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grob T, Al Hinai ASA, Sanders MA, Kavelaars FG, Rijken M, Gradowska PL, et al. Molecular characterization of mutant TP53 acute myeloid leukemia and high-risk myelodysplastic syndrome. Blood. 2022;139:2347–54. doi: 10.1182/blood.2021014472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schimmer RR, Kovtonyuk LV, Klemm N, Fullin J, Stolz SM, Mueller J, et al. TP53 mutations confer resistance to hypomethylating agents and BCL-2 inhibition in myeloid neoplasms. Blood Adv. 2022;6:3201–6. doi: 10.1182/bloodadvances.2021005859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gurney M, Mangaonkar AA, Lasho T, Finke C, Al-Kali A, Gangat N, et al. Somatic TP53 single nucleotide variants, indels and copy number alterations in chronic myelomonocytic leukemia (CMML) Leukemia. 2023;37:1753–6. doi: 10.1038/s41375-023-01964-3. [DOI] [PubMed] [Google Scholar]
- 5.AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 2017;7:818-31. [DOI] [PMC free article] [PubMed]
- 6.Hafner A, Bulyk ML, Jambhekar A, Lahav G. The multiple mechanisms that regulate p53 activity and cell fate. Nat Rev Mol Cell Biol. 2019;20:199–210. doi: 10.1038/s41580-019-0110-x. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y-H, Yao C-Y, Lin C-C, Gurashi K, Amaral FMR, Bossenbroek H, et al. A three-gene leukaemic stem cell signature score is robustly prognostic in chronic myelomonocytic leukaemia. Br J Haematol. 2023;201:302–7. doi: 10.1111/bjh.18681. [DOI] [PubMed] [Google Scholar]
- 8.Hurtado AM, Luengo-Gil G, Chen-Liang TH, Amaral F, Batta K, Palomo L, et al. Transcriptomic rationale for synthetic lethality-targeting ERCC1 and CDKN1A in chronic myelomonocytic leukaemia. Br J Haematol. 2018;182:373–83. doi: 10.1111/bjh.15408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Unnikrishnan A, Papaemmanuil E, Beck D, Deshpande NP, Verma A, Kumari A, et al. Integrative genomics identifies the molecular basis of resistance to Azacitidine therapy in myelodysplastic syndromes. Cell Rep. 2017;20:572–85. doi: 10.1016/j.celrep.2017.06.067. [DOI] [PubMed] [Google Scholar]
- 10.Shi D, Jiang P. A different facet of p53 Function: regulation of immunity and inflammation during tumor development. Front Cell Dev Biol. 2021;9:762651. [DOI] [PMC free article] [PubMed]
- 11.Patnaik MM, Tefferi A. Chronic myelomonocytic leukemia: 2024 update on diagnosis, risk stratification and management. Am J Hematol. 2024;99:1142−65. [DOI] [PMC free article] [PubMed]
- 12.Sallman DA, Borate U, Cull EH, Donnellan WB, Komrokji RS, Steidl UG, et al. Phase 1/1b study of the stapled peptide ALRN-6924, a dual inhibitor of MDMX and MDM2, as monotherapy or in combination with cytarabine for the treatment of relapsed/refractory AML and advanced MDS with TP53 Wild-Type. Blood. 2018;132:4066. doi: 10.1182/blood-2018-99-118780. [DOI] [Google Scholar]
- 13.Rodriguez-Meira A, Norfo R, Wen S, Chédeville AL, Rahman H, O’Sullivan J, et al. Single-cell multi-omics identifies chronic inflammation as a driver of TP53-mutant leukemic evolution. Nat Genet. 2023;55:1531–41. doi: 10.1038/s41588-023-01480-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Kong G, Rajagopalan A, Lu L, Song J, Hussaini M, et al. p53−/− synergizes with enhanced NrasG12D signaling to transform megakaryocyte-erythroid progenitors in acute myeloid leukemia. Blood. 2017;129:358–70. doi: 10.1182/blood-2016-06-719237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, Sun Q. TP53 mutations, expression and interaction networks in human cancers. Oncotarget. 2017;8:624–43. doi: 10.18632/oncotarget.13483. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this article will be provided to collaborating investigators through reasonable request to the corresponding authors after requisite institutional review board approval. The data are not publicly available due to privacy or ethical restrictions.