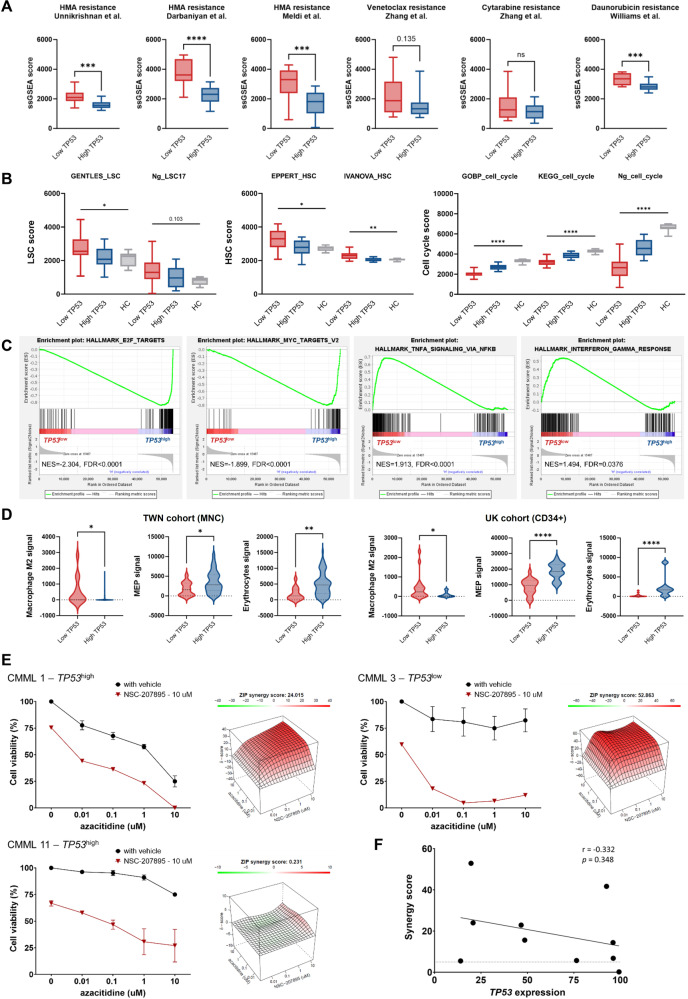
Fig. 2. Biological and therapeutic implications of TP53 expression in CMML.
A Box plots displaying resistance signatures derived from single-sample GSEA for hypomethylating agents (HMA), venetoclax, cytarabine, and daunorubicin in patients with lower and higher TP53 expression in the UK CD34 + -sorted cohort. B Box plots displaying scores of leukemic stem cell (LSC), hematopoietic stem cell (HSC), and cell cycle of patients with TP53high and TP53low expression and healthy controls (HC) in the UK CD34 + -sorted cohort. C Representative GSEA plots of pathway enrichment in CMML patients with the lowest 25% vs highest 25% TP53 expression in the UK CD34 + -sorted cohort. D Violin plots displaying different signatures seen in the Taiwan discovery cohort and the UK validation cohort. MEP: megakaryocytic-erythroid progenitors. A, B, D ****P ≤ 0.0001, ***P ≤ 0.001, **P ≤ 0.01, *P ≤ 0.05. P values were computed using Mann–Whitney or Kruskal–Wallis test. E Representative 3D synergy plots using zero interaction (ZIP) model (right) and dose response curves (left) for CMML bone marrow mononuclear cells (n = 3 patients; mean + SEM) treated for 72 h ex vivo with NSC-207895 and azacitidine combination at various concentrations. The presence of synergy was determined utilizing the SynergyFinder computational package and the ZIP synergy index where red denotes synergism and green denotes antagonism. A positive synergy score is the percent more cell death than expected. F Dot plot displaying the correlation between TP53 expression and synergy score of 10 patient samples.