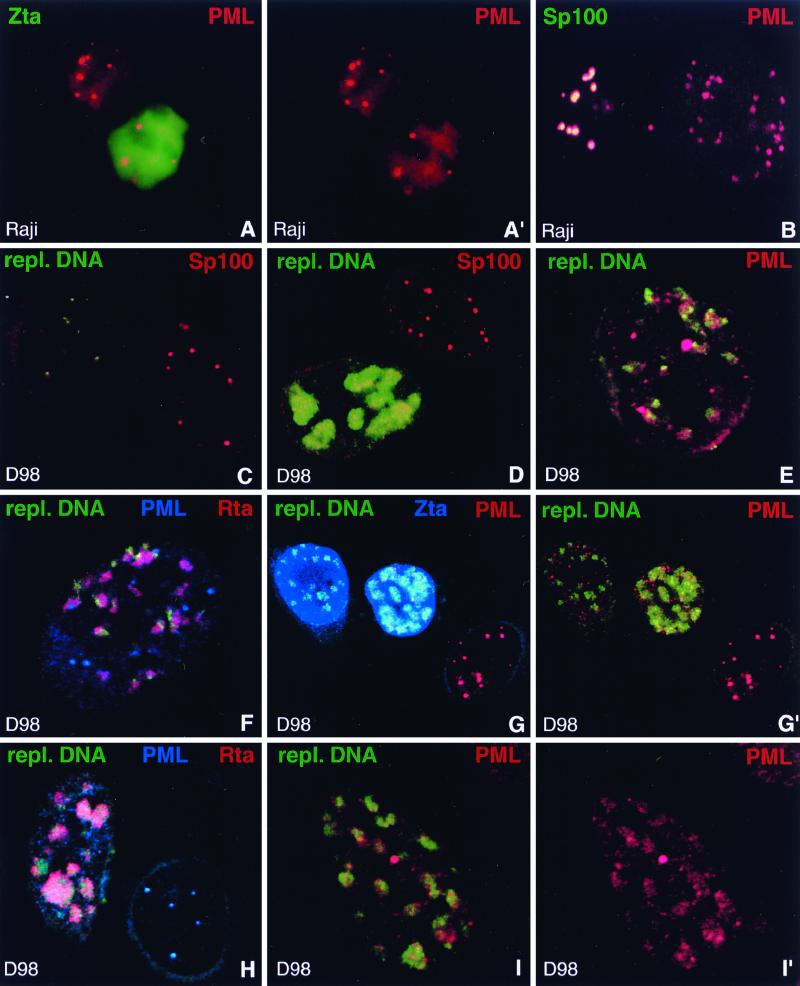
FIG. 4.
Behavior of PML upon EBV reactivation compared to Sp100. (A and A′) Raji cells stained for Zta and PML after induction of lytic activation. Unlike other ND10 proteins, PML often remains concentrated in ND10 in Zta-positive cells. A′, PML staining only. (B) Raji cells double labeled with antibodies against PML and Sp100 after disruption of latency to demonstrate release of Sp100 but not of PML from ND10. The left-hand cell shows typical ND10 which contain both proteins, indicating that EBV remained latent. In the right-hand cell, EBV apparently entered the lytic cycle and released Sp100 from ND10 which still contain PML. (C) D98/HR1 cells were Zta transfected to induce lytic activation, stained for Sp100, and probed with a labeled PCR product which recognizes clusters of replicating EBV DNA but not individual episomes. Note the lack of detectable hybridization signal in the right-hand cell, in which no lytic activation occurred. This cell contains Sp100-positive ND10. In contrast, the left-hand cell displays small, i.e., early, foci of replicating EBV DNA. Sp100 is no longer concentrated in ND10 as soon as replicated EBV genomes are detectable. (D) Zta-transfected D98/HR1 cells labeled for Sp100 and hybridized with the probe specific for replicating EBV DNA. The lower cell shows an advanced stage of the lytic cycle, with replicating EBV genomes that form large clusters of DNA. Sp100 remains dispersed and does not associate with the replication domains. In the upper cell, no lytic cycle has been induced and Sp100 is not dispersed from ND10. No replicated EBV DNA is detectable in this cell. (E) A Zta-transfected D98/HR1 cell was stained for PML and probed for replicating EBV DNA. In contrast to Sp100, PML-positive ND10 are still visible after lytic replication has started. Most of the replication domains are found in close association with PML. (F) D98/HR1 cells were transfected with Zta to induce lytic activation and triple labeled for PML and Rta by immunostaining and for replicating EBV genomes by FISH. Rta forms aggregates in close association with the replication domains but without complete colocalization. PML is still visible in the form of several ND10-like structures. (G and G′) Zta-transfected D98/HR1 cells triple labeled for the detection of PML, Zta, and replicating EBV DNA. The replication-specific probe detects EBV genomes only in Zta-positive cells (two leftmost cells). In the right-hand untransfected cell, PML is almost completely aggregated into ND10, whereas PML becomes increasingly dispersed during lytic replication (two leftmost cells). However, even during late replication stages, as represented by the middle cell, PML is still concentrated in several ND10-like foci. In panel G′, Zta staining is omitted. (H) Zta-transfected D98/HR1 cell immunostained for Rta and PML and probed for replicating EBV DNA. In the right-hand cell, which is probably not transfected, no lytic activation was induced. The replication-specific probe fails to recognize EBV-specific genomes in this cell, which displays intact PML-positive ND10. The left-hand cell represents a late stage of the lytic cycle, as judged from the size of the replication compartments. PML foci have almost completely disappeared. In contrast to earlier replication stages as shown in panel F, the Rta aggregations and replication domains show a higher degree of colocalization, suggesting that both components had merged while increasing in size. (I and I′) Zta-transfected D98/HR1 cell hybridized with the replication-specific probe and immunostained for PML. PML is redistributed into the replication domains and colocalizes almost completely with the replicated EBV genomes in the form of diffuse PML accumulations. I′, PML staining only.