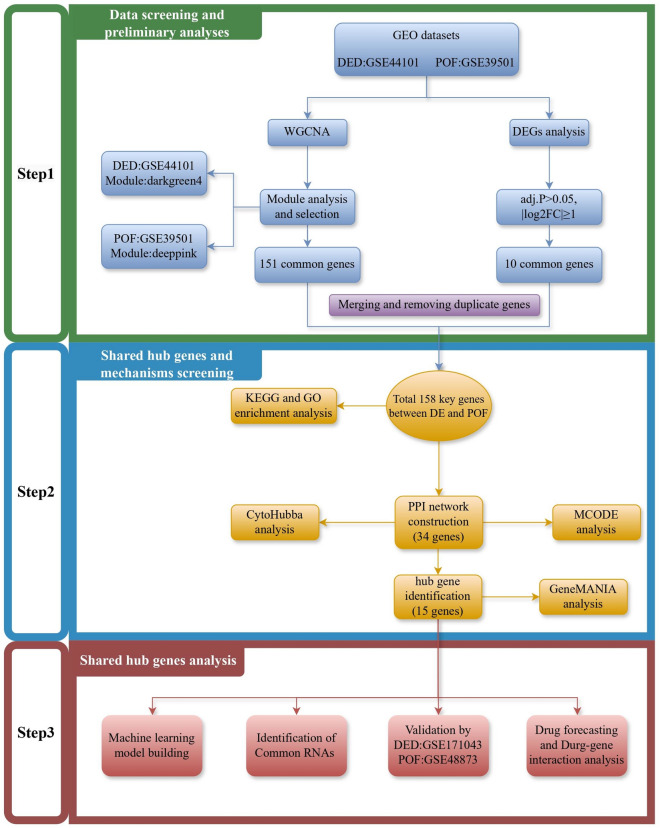
Figure 1.
Schematic diagram of the general flow of this research work. In order to gain a more penetrating understanding of the mechanisms of comorbidity between POF and DED, we began our research journey using data from the GEO database. Specifically, this study was divided into three main parts. Firstly, in the first part we identified the GSE44101 and GSE39501 datasets as the primary cohorts through a careful matching strategy, and identified the GSE171043 and GSE48873 datasets for validation. We performed WGCNA analysis on GSE44101 and GSE39501 to identify the key modules and module genes, followed by differential expression analysis, respectively. The 158 common genes of POF and DED were obtained by merging and removing duplicate genes. Subsequently, in the second part, in order to reveal the functional basis of these common key genes, we performed exhaustive GO and KEGG analyses. Meanwhile, in order to further screen the hub genes, we executed a set of step-by-step screening of 15 hub genes after covering PPI, MCODE, Cytoscape, and GeneMANIA analyses. In the third part, the hub genes were further analysed and validated through seamless integration with 4 different methods and 2 datasets. This comprehensive approach not only enriches our understanding of the mechanisms of POF and DED comorbidity, but also lays a solid foundation for identifying prospective targeted therapies and interventions for the treatment of POF and DED comorbidity.