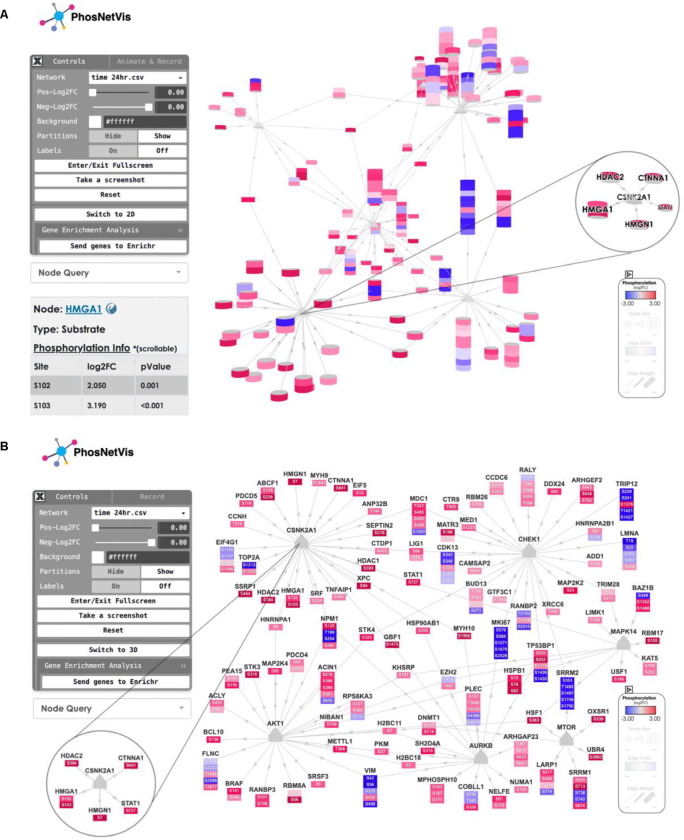
Figure 4. Snapshots of network visualization interface featuring the phosphorylation landscape of SARS-CoV-2 infection.
This page allows users to view and interact with the network curated from the Global Phosphorylation Landscape of SARS-CoV-2 Infection (Bouhaddou et al., 2020). Users can rotate the network, zoom in/out, pan through, see a node information by double clicking on it, or reset the view by double clicking on the background. Additionally, it provides options for the user to switch between 3D or 2D network. Users can also send their genes list to the Enrichr tool23–25 for gene enrichment analyses, for further exploration of enriched pathways and functional gene groups. Double-clicking on any node opens an interactive table that displays detailed information on the node as shown in Panel A bottom left for node HMGA1. A) Snapshot of the 3D view of the network, including a magnified view of a CSNK2A1-central subnetwork in 3D. This subnetwork highlights phosphorylation sites known to be associated with cytoskeleton remodeling, filtered from the main network. B) Snapshot of the 2D view of the same network. In each panel, the left corner shows the control panel; the middle displays the network; the bottom right corner contains the legend. The starred section indicates the CSNK2A1 phosphorylation subnetwork, including a magnified view of the same CSNK2A1 node with select interactors in a subnetwork in 2D. These features enable comprehensive exploration and analysis of the phosphorylation events during SARS-CoV-2 infection.