Abstract
Low pH stress and its influence on antibody binding is a common consideration among chemists, but is only recently emerging as a consideration in Immunological studies. Antibody characterizations in Multiple Sclerosis (MS), an autoimmune disease of the Central Nervous System (CNS) has revealed that antibodies in the cerebrospinal fluid (CSF) of patients with Multiple Sclerosis bind to myelin-related and non-myelin antigen targets. Many laboratories have used molecular biology techniques to generate recombinant human antibodies (rhAbs) expressed by individual B cells from healthy donors and patients with systemic autoimmune disease to identify antigen targets. This approach has been adapted within the Neuroimmunology research community to investigate antigen targets of individual B cells in the CSF of MS patients. Our laboratory determines which antibodies to clone based on their immunogenetics and this method enriches for cloning of rhAbs that bind to neurons. However, newer technologies to assist in purification of these rhAbs from culture supernatants use an acidic elution buffer which may enhance low pH stress on the antibody structure. Our laboratory routinely uses a basic elution buffer to purify rhAbs from culture supernatants to avoid low pH stress to the antibody structure. Our goal was to investigate whether acidic elution of our rhAbs using Next Generation Chromatography would impact the rhAbs’ ability to bind neurons. The limited data presented here for two neuron-binding rhAbs tested indicated that acidic elution buffers used during rhAb purification impacted the ability of rhAbs with low CDR3 charge to maintain binding to neuronal targets. Reproducibility in a larger panel of rhAbs and factors underlying these observations remain untested.
Keywords: Neuronal binding, Antibody isolation, Antibody antigen interaction, CDR3 charge
1. Introduction
Multiple Sclerosis (MS) is an autoimmune disease of the central nervous system that typically manifests in the 2nd, 3rd or 4th decades of life.(Lublin et al., 2014; Steinman, 2014; Friese et al., 2014; Hauser et al., 2013) Diversity of 1) clinical presentations, 2) lesion pathology and 3) autoimmune responses exhibited by these patients can impact the quality of patient care, as treatment benefit is likely dependent on the nature of an individual patient’s disease mechanism. Thus, understanding the pathways of MS evolution can inform us regarding whether early treatment will impact an individual patient’s disease course.
Multiple, heterogeneous pathogenic mechanisms are likely to play a role in MS. However, the impressive efficacy of B cell depletion therapy (BCDT) in MS (with nearly 50% experiencing no evidence of disease activity),(Calabresi, 2017) indicates a likely role of B cells in the pathology of MS. BCDT does not target antibody producing B cells, suggesting that antigen presentation or cytokine production by B cells are likely mechanisms of B cell involvement in MS autoimmunity. Of note, both antigen presentation and cytokine output by a B cell requires that its membrane-bound antibody binds to its cognate antigen and internalizes the antibody-antigen complex. Thus, identifying the antigen specificity of antibodies expressed by B cells in patients with Multiple Sclerosis is critical to understanding of humoral autoimmune mechanisms.
Antibodies expressed by CD19+ B cells in the CSF of MS patients are either reactive to myelin components(von Budingen et al., 2008) or do not bind brain tissue.(Owens et al., 2009) In contrast, we have documented that 93.75% (30 out of 32) of CD19+ B cells (including PBs) that we isolated from the CSF of 10 treatment-naive early MS patients produce antibodies that react to cytosolic targets in neurons.(Ligocki et al., 2015) These antibodies were generated by isolating the antibody heavy and light chain sequences from individual B cells, cloning them into expression vectors and using HEK293 cells to produce the recombinant human antibodies into the cultures. Isolation of the recombinant human antibodies (rhAbs) from culture supernatants was done using Protein G gravity columns, which we identified as a labor-intensive step in the process. Thus, we acquired a Next Generation Chromatography (NGC) instrument to facilitate efficiency in isolating rhAbs and thus expand the number of rhAbs we could test in a period of time.
The recommended protocol on the NGC utilized an acidic elution buffer. We had routinely used a basic elution buffer as low pH conditions can impact both constant domain and variable domain structure.(Zhou et al., 2018; Imamura, 2019) For example, low pH conditions can lead to unfolding and aggregation of IgG1 and IgG2 antibody molecules,(Mazzer et al., 2015; Lopez et al., 2019) completely collapse the second constant domain,(Latypov et al., 2012) and permit further flexibility in the CH2 that allows for the variable domain to fold over onto the constant domain.(Gagnon et al., 2015) Variable domain antigen binding can also be impacted, but pH-dependent binding is highly variable for reasons that remain to be fully elucidated(Zhou et al., 2018; Latypov et al., 2012; Gagnon et al., 2015) and can even lead to polyreactivity.(Djoumerska-Alexieva et al., 2009; McMahon and O’Kennedy, 2000) There is also evidence that interchain disulfide bonds at particular residues in the constant domain can impact variable domain stability.(Rothlisberger et al., 2005) Of note, these previous studies typically incubated the antibody molecule(s) in low pH conditions for extended periods of time whereas the elution protocol recommended to us included an immediate desalting step following elution to return the antibody preparation to neutral pH prior to fraction collection. To test whether acidic elution would impact neuron binding, we identified two rhAbs that we had previously documented bound neurons and compared neuronal reactivity of these two rhAbs prepared by the NGC-recommended protocol and our protocol. The results indicate that neuron binding by one of these two rhAbs was compromised when using acidic elution during the purification process.
2. Methods
2.1. Patient subjects
Antibody heavy and light chain gene pairs are obtained from single sorted B cells found in the cerebrospinal fluid and blood of patients with Multiple Sclerosis.(Ligocki et al., 2015; Rivas et al., 2017) There are 767 heavy chains included in this analysis with 9 patients represented in this dataset. There are 146 recombinant human antibodies (rhAbs) included in this analysis with three featured recombinant human antibodies (JRR67, JRR28 and JRR44) obtained from 3 different MS patients. Of note, JRR67 does not bind to neurons in mouse brain tissue whereas JRR28 and JRR44 bind to neurons in mouse brain tissue when eluted with a basic elution buffer.(Rivas et al., 2017)
2.2. Recombinant human antibody generation and isolation by next generation chromatography
HEK293T cells (ATCC, Manassas, VA) were maintained in HyClone Dulbeccos Modified Eagles Medium (DMEM) (GE Healthcare Life Sciences). All recombinant human antibodies (rhAbs) were transiently transfected into HEK293T cells with the lipid transfection reagent JetPEI (PolyPlus Transfection) as done previously.(Ligocki et al., 2015; Rivas et al., 2017) Supernatants from these cultures were collected on days 3, 5, 7 and 10. The cell pellets were spun down and supernatants were passed through 0.2um filters and subjected to antibody purification on the NGC QUEST 10 system. The flow rate of the culture supernatant into the system was controlled by Chromelab v6 software. Samples first pass through a Bio-Scale Mini UNOsphere SUPrA affinity chromatography column (BioRad), eluted with either acidic buffer (100 mM glycine, pH = 2.5) or basic buffer (50 mM diethylamine, 150 mM NaCl, pH = 12), followed by 10 mL Bio-Scale Mini Bio-Gel P-6 desalting column (BioRad) and collected into fractions. Purified antibody in each fraction was detected by UV light. The chromatography plots of the acidic and basic elutions of the three rhAbs used as examples are presented in Supplemental Fig. 1. Each of the three rhAbs were eluted two separate times in either the acidic or basic elution buffers. Data in the figures is representative of the second preparation of each rhAb in acidic or basic elution buffer. The concentrations of the antibodies were determined by Sandwich ELISA as done previously.(Ligocki et al., 2015; Rivas et al., 2017)
2.3. Immunocytochemistry of SH-SY5Y cells
SH-Sy5y cells were purchased from ATCC (Cat No. CRL-2266, Manassas, VA) and cultured in media containing eagle’s minimum essential medium and F12 medium in 1:1 mixture supplemented with 10% heat-inactivated fetal bovine serum (Gibco) and 1% penicillin-streptomycin (Gibco). Cells were passaged every 4–7 days after attaining over 80% confluency. For immunocytochemistry experiments with SH-Sy5y cells, glass coverslips (Fisher Scientific) were coated with laminin (Sigma-Aldrich) at 50 μg/mL in dPBS for 2 h at 37 °C, washed once with PBS and SH-Sy5y cells were plated for 48 h. Cells were washed with PBS, fixed with 4% PFA, washed again and permeabilized with PBS + 0.2% Triton X − 100 (Sigma) + 2 mg/mL BSA (Sigma) in PBS for 10 min, then washed in PBS. Cells were then blocked with PBS+ 1% goat serum (Life Technologies), 3% BSA and 0.1% Triton X-100 for 2 h at room temperature before adding primary rhAbs at 20 μg/mL in blocking buffer overnight at 4 °C. The next day, coverslips were rinsed four times with PBS + 1% Goat Serum +0.025% Triton X-100 for 3 min, then incubated with secondary goat anti-human secondary antibody conjugated to AlexaFluor 488 (Invitrogen) diluted in blocking buffer at a factor of 1:1000 for 1 h at room temperature. The rinses were repeated, and the cells were counterstained with DAPI (Sigma) in PBS for 5 min. Coverslips were mounted onto the glass slides using FluoroMount G (SouthernBiotech). Slides were visualized using a Zeiss LSM780 confocal microscope with a 20×/0.80 NA air immersion objective. These experiments were repeated twice.
2.4. Immunofluorescence imaging of rhAb binding to mouse cortical cells
Primary mouse cortical cells (PMCC) were isolated from postnatal day 0–2 C57Bl/6 J pups and digested using 0.25% Trypsin-EDTA to isolate cells as previously described.(Ortega et al., 2020; Ujas et al., 2023) Briefly, PMCC were plated at 250,000 cells/well of a 48-well plate (Corning Primaria) in PMCC culture media. A complete media change was done at day-in-vitro (DIV) 3 and cells were cultured at 37 °C with 5% CO2 until confluent (minimum DIV7) prior to experimental use. These cultures result in a mixture of neurons (~9.5%), astrocytes (~45.7%) and microglia (~7.9%)(Supplemental Fig. 2). For PMCC staining, cells were fixed with 4% PFA, washed and permeabilized with 0.2% Triton X-100 in 2 mg/mL BSA. Cells were then blocked with PBS + 1% goat serum, 3% BSA and 0.1% Triton X-100 for 2 h at room temperature before adding primary rhAbs at 20 μg/mL in blocking buffer overnight at 4 °C. The next day, coverslips were rinsed four times with PBS + 1% Goat Serum +0.025% Triton X-100 for 3 min, then incubated with secondary goat anti-human secondary antibody conjugated to AlexaFluor 488 diluted in blocking buffer at a factor of 1:1000 for 1 h at room temperature. The rinses were repeated, and the cells were counterstained with DAPI in PBS for 5 min. Coverslips were mounted on glass slides using FluoroMount G. Slides were visualized using a Zeiss LSM780 confocal microscope with a 20×/0.80 NA air immersion objective. These experiments were repeated twice.
2.5. Quantification of fluorescent intensity
Images of stained cells were analyzed using Fiji software.(Schindelin et al., 2012) Images were captured at 20× magnification at the same exposure times for qualitative comparison across different antibodies. Images were then analyzed on Fiji (ImageJ, V 1.52). Given the heterogeneity in cell cultures and images, we used the region of interest (ROI) feature to outline areas with cells in each image. Then, fluorescent intensity of all ROIs was measured using integrated density (IntDen) values. The background IntDen for regions without cells was subtracted to obtain net fluorescence intensity. The values were then averaged per pixel area and compared across different treatments as noted in the figures.
2.6. Binding of rhAbs by ELISA
Lysate was made from human spinal cord as previously described. (Rivas et al., 2017) Plates were coated with 10 μg/mL of lysate in bicarbonate buffer overnight at 4 °C. The next day, plates were washed twice with PBST and blocked with 3% BSA in PBST for 2 h at room temperature. Plates were washed twice and rhAbs was added at 10, 5, and 1 μg/mL in 1% BSA and incubated overnight at 4 °C. The following day, plates were washed three times followed by incubation with 1 μg/mL of biotinylated anti-human IgG (eBioscience, San Diego, CA) in 1% BSA was incubated for 2 h at room temperature. Plates were washed three more times, followed by incubation with a 1:2000 dilution of streptavidin-HRP (BD Pharmigen, San Jose, CA) in 1% BSA for 1 h at room temperature. Plates were washed three times, developed for 30 s with TMB substrate (eBioscience, San Diego, CA), and neutralized with 1 M HCl before reading at 450 nm with an Epoch Microplate Spectrophotometer (BioTek, Winooski, VT). This experiment was repeated twice with two different preparations of rhAb eluted in basic and acidic buffer.
2.7. CDR3 charge calculation
CDR3 charge calculations were performed on the VDJServer web portal(Christley et al., 2018) using the Alakazam program.(Gupta et al., 2015) The conserved residues were trimmed from the sequence when doing the calculation. CDR3 charge was calculated as difference between the number of positively charged amino acids (arginine, Arg, R and lysine, Lys, K) and negatively charged amino acids (aspartic acid, Asp, D and glutamic acid, Glu, E).
3. Results and discussion
Recombinant human full length IgG1 antibody technology was pioneered in the early 2000’s with the intention of identifying the prevalence of self-reactive B cells at distinct stages of development in healthy individuals.(Wardemann et al., 2003) This technology was thereafter applied to antibodies expressed by B cells in the setting of autoimmune disease, to identify antigen targets that might infer pathological involvement. In Multiple Sclerosis specifically, this technology was used by 3 laboratories(von Budingen et al., 2008; Owens et al., 2009; Ligocki et al., 2015) to produce 92 rhAbs from antibody secreting B cells in the cerebrospinal fluid or brain tissue of 23 patients and test for neuronal binding on human brain tissue. Only our laboratory demonstrated significant binding to neurons in brain tissue (92% of the 32 rhAbs tested), which we attributed to our use of a gentle fixation approach (4% PFA) to preserve antigen integrity.(Lublin et al., 2014) However, others using gentle fixation were unable to demonstrate that rhAbs produced in their laboratory significantly bound to neurons.(von Budingen et al., 2008; Blauth et al., 2015)
A second variable among the 3 laboratories was the method of antibody elution from Protein A/G columns. Our laboratory had been using a basic elution buffer to avoid low pH stress on the antibody eluate which can cause aggregation or compromise the ligand binding properties.(Zhou et al., 2018; Imamura, 2019) Careful examination of the elution methods of rhAbs used by other laboratories indicated that the other 2 laboratories had used an acidic elution buffer to isolate antibodies from culture supernatants. All 3 laboratories performed extensive buffer exchange to return the rhAb solution to a neutral pH following elution. Yet, we could not rule out that their conclusions could be based on an enzymatic issue related to the elution step. Upon acquisition of Next Generation Chromatography instrumentation, we asked whether the recommended acidic elution buffer to purify rhAbs would impact antibody reactivity. To do this, we chose 3 rhAbs from our portfolio: one rhAb (JRR67), which does not bind neurons in mouse brain tissue, and two rhAbs (JRR28 and JRR44) which bind neurons in mouse brain tissue.(Rivas et al., 2017) We first tested the ability of these three rhAbs to bind SH-Sy5y cells, which are a human neuroblastoma cell line (Fig. 1). JRR67 maintained negative binding to SH-Sy5y when eluted with the acidic buffer (Panel A). JRR28 maintained positive binding to SH-Sy5y when eluted with the acidic buffer (Panel B). JRR44 no longer bound SH-Sy5y when eluted with the acidic buffer (Panel C). Binding of these three rhAbs when eluted with basic buffer are provided in Panels D-F for comparison, and indicated that there was a 4.6-fold reduction in JRR44 binding to SH-Sy5y when the rhAb was eluted with the acidic buffer.
Fig. 1.
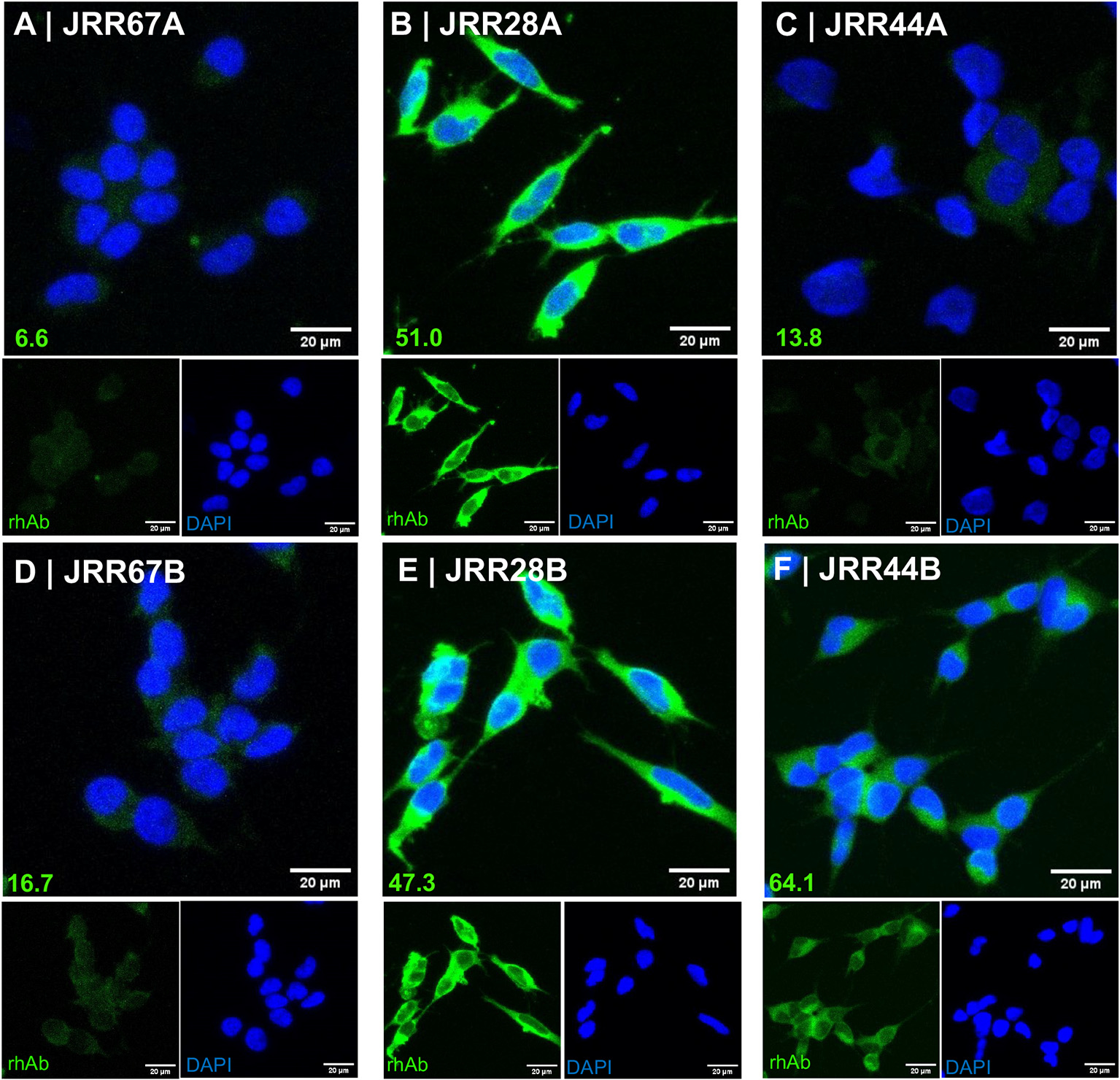
JRR44 no longer binds SH-Sy5y cells when isolated by acidic elution. (A-C) Immunocytochemistry of SH-Sy5y cells stained with JRR67 (A), JRR28 (B), and JRR44 (C) isolated with an acidic elution buffer. (D-F) Immunocytochemistry of SH-Sy5y cells stained with JRR67 (D), JRR28 (E), JRR44 (F) isolated with a basic elution buffer. Normalized values of fluorescence intensity are at the bottom left of each panel and scale bars at the bottom right of each panel.
The second test utilized immunofluorescence of primary mouse cortical cells, which are a mixture of neurons, astrocytes and microglia (Ortega et al., 2020; Ujas et al., 2023) (Fig. 2). JRR67 maintained negative binding to mouse cortical cells when eluted with the acidic buffer (Panel A). JRR28 maintained positive binding to mouse cortical cells when eluted with the acidic buffer (Panel B). JRR44 no longer bound mouse cortical cells when eluted with the acidic buffer (Panel C). Binding of these three rhAbs when eluted with basic buffer are provided in Panels D-F for comparison, and indicated that there was a complete abrogation in JRR44 binding to primary mouse cortical cells when the rhAb was eluted with the acidic buffer.
Fig. 2.
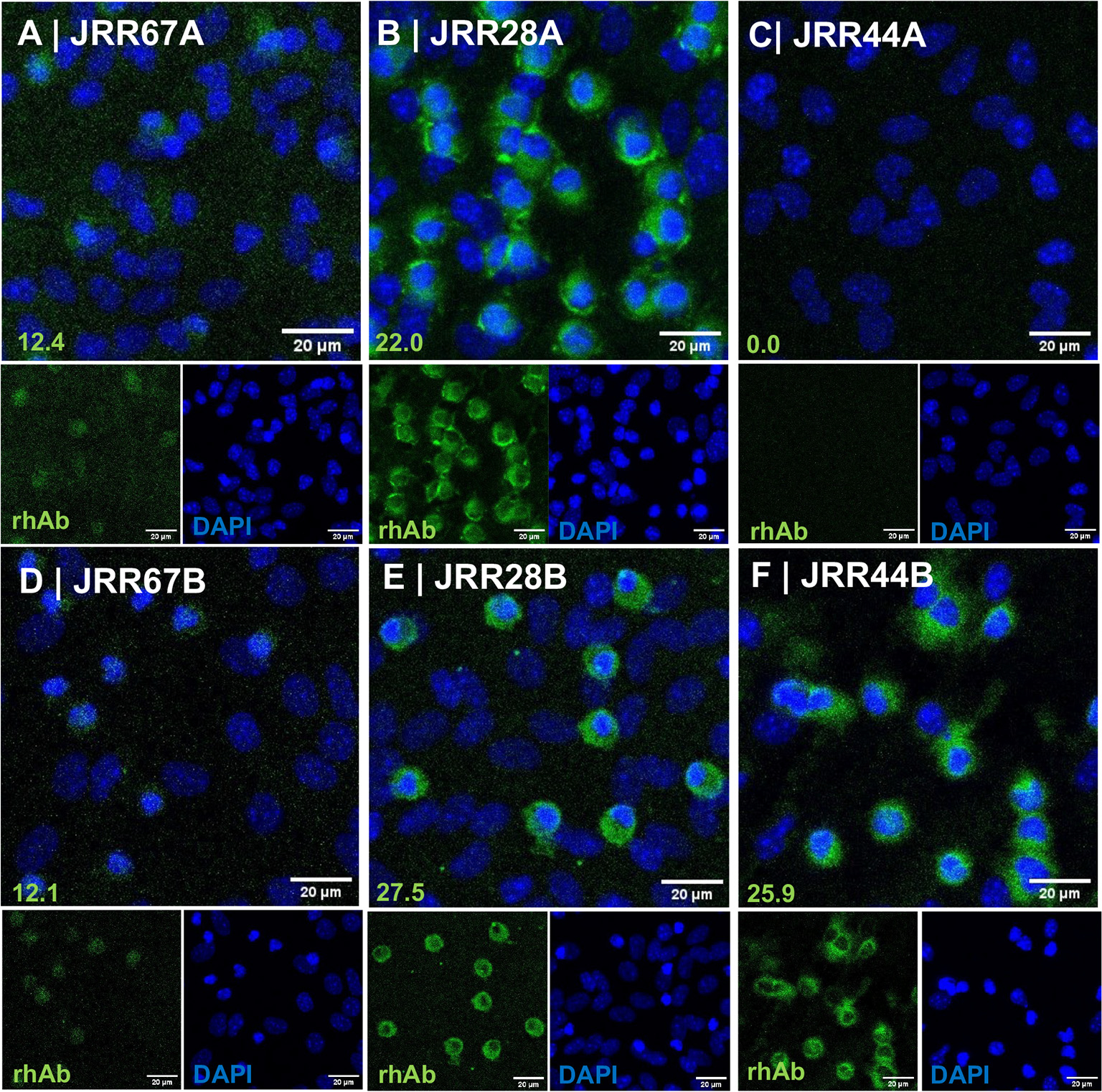
JRR44 no longer binds primary mouse cortical cells when isolated by acidic elution. (A-C) Immunocytochemistry of primary cortical cells stained with JRR67 (A), JRR28 (B), and JRR44 (C) isolated with an acidic elution buffer. (D-F) Immunocytochemistry of primary cortical cells stained with JRR67 (D), JRR28 (E), JRR44 (F) isolated with a basic elution buffer. Normalized values of fluorescence intensity are at the bottom left of each panel and scale bars at the bottom right of each panel.
The third test utilized ELISA of human spinal cord lysates as a target pool since the rhAbs we were testing came from patients whose disease manifested initially in the spinal cord (Fig. 3).(Rivas et al., 2017) Spinal cord lysate was used to address potential affinity affects. JRR67 maintained negative binding to human spinal cord lysate when eluted with the acidic buffer compared to basic buffer (Left Panel). JRR28, maintained positive binding to SH-Sy5y when eluted with the acidic buffer compared to basic buffer (Middle Panel). JRR44 no longer bound human spinal cord lysate when eluted with the acidic buffer even at the highest concentration of lysate in the assay (Right Panel).
Fig. 3.
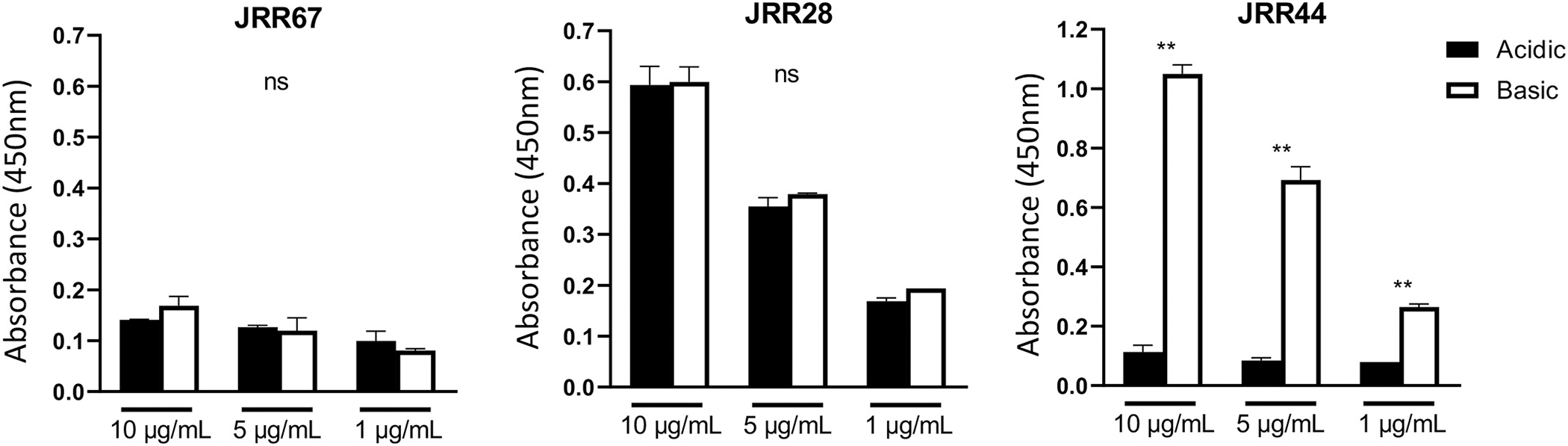
JRR44 no longer binds human spinal cord lysate when isolated by acidic elution. Human spinal cord lysate was used as a binding target for rhAbs JRR67, JRR28 and JRR44 that were isolated either by acidic (black bars) or basic elution (white bars). Three dilutions of rhabs (10μg/ml, 5μg/ml and 1μg/ml) were used. Y-axis indicates the absorbance at 450 nm. ** p < 0.01, ns not significant.
Others had suggested that the immunogenetics of the antibody may influence susceptibility to low pH stress.(Zhou et al., 2018) Comparison of the antibody variable heavy chain gene characteristics of JRR28 and JRR44 indicated that JRR44, had a CDR3 charge of − 4 and a variable heavy chain gene isoelectric point of 4.97 (Table 1). The isoelectric point of the antibody variable heavy chain reflected that (pI = 4.97).
Table 1.
rhAb immunogenetics information.
| rhab name | JRR28 | JRR44 | JRR67 |
|---|---|---|---|
|
| |||
| VH gene | IGHV4–59 | IGHV4–59 | IGHV4–30 |
| VL gene | IGLV3–11 | IGLV2–8 | IGKV2–30 |
| J gene usage | IGHJ5 | IGHJ6 | IGHJ5 |
| Concentration after BASIC Elution 1 | 1.48 | 1.82 | 2.70 |
| Concentration after ACIDIC elution 1 | 1.37 | 1.79 | 2.75 |
| charge of CDR3 | −1 | −4 | 0 |
| length of CDR3 | 22 | 20 | 19 |
| mutation frequencies | 7 | 15 | 11 |
| heavy chain pI | 7.77 | 4.97 | 9 |
Concentration is expressed in micrograms per original milliliter of culture supernatant.
Next, we asked how frequently the charge of antibody heavy chain CDR3 regions that we had cloned were outside the mean (Fig. 4). We found that of the 146 antibodies we had cloned, 6 of them (4.1%) displayed antibody heavy chain CDR3 charges of − 3 or − 4 (Panel A). The total antibody candidate pool (n = 767) from which we selected the antibodies to clone had 44 antibodies (5.7%) that displayed antibody heavy chain CDR3 charges of − 3 or − 4 (Panel B). The median antibody heavy chain CDR3 charge for both the cloned antibodies and the antibody candidate pool was − 0.01. We did not have access to the antibody variable heavy chain genes of rhAbs produced by our neuroimmunology colleagues from B cells in the CSF or brain of MS patients,(von Budingen et al., 2008; Owens et al., 2009; Blauth et al., 2015; Willis et al., 2015; Yu et al., 2011; Zhang et al., 2005) but it would be interesting to determine how many of the rhAbs characterized to date may have been susceptible to low pH stress during column elution.
Fig. 4.
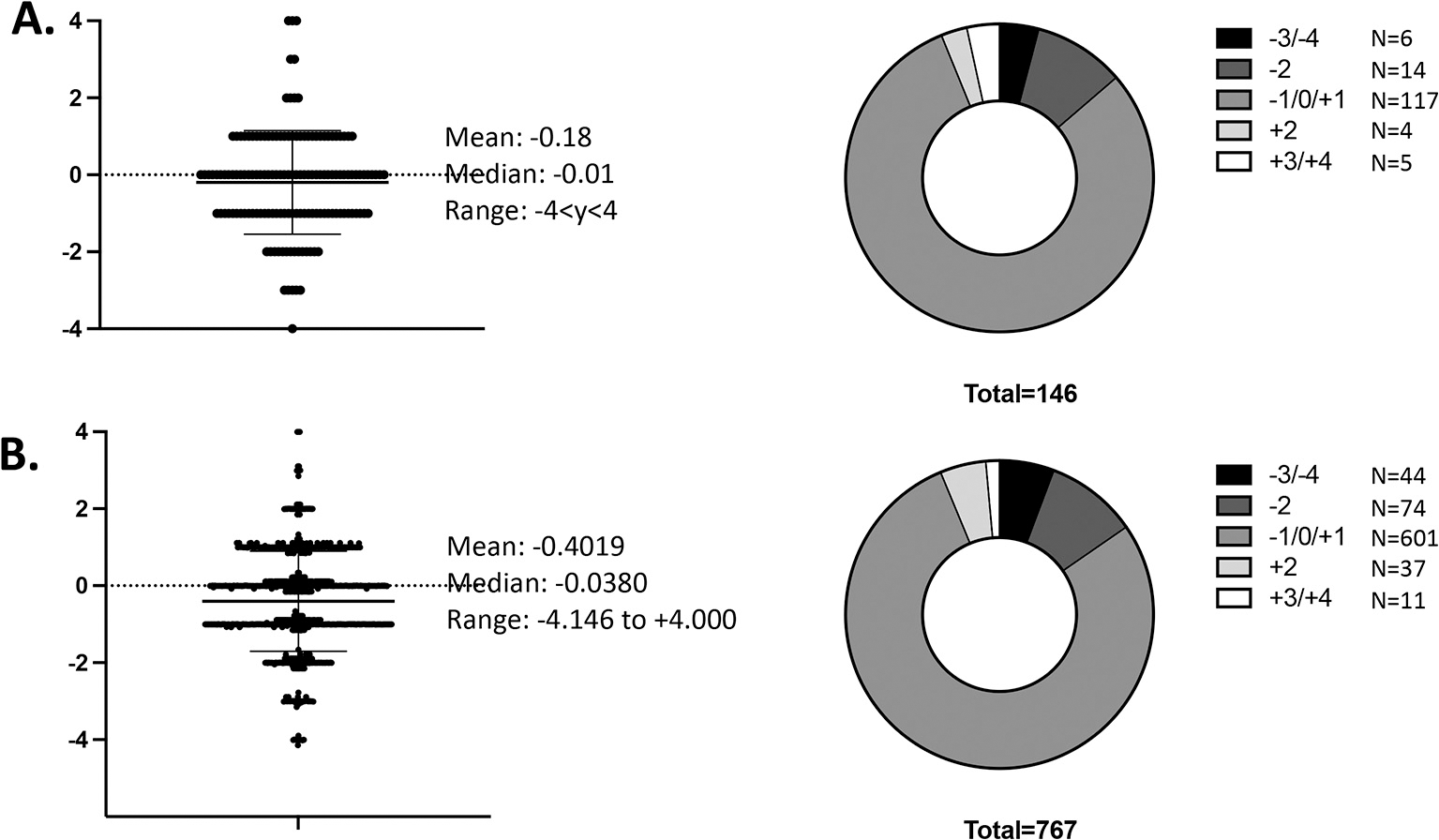
Patients with Multiple Sclerosis harbor antibodies with highly negative CDR3 charges. (A) Portfolio of recombinant human antibodies (rhAbs) that have been expressed and tested for binding to human brain tissue. (B) Portfolio of all rhAbs in queue to be expressed and tested for binding to human brain tissue.
4. Conclusion
We used three distinct methods to test the influence of low pH stress on neuronal binding by rhAbs: 1) immunocytochemistry (ICC) of the human neuroblastoma cell line, SH-Sy5y (Fig. 1), 2) immunofluorescence cytochemistry (IFC) using mouse cortical cells (Fig. 2) and 3) Enzyme-linked Immuno Assay (ELISA) of human spinal cord lysate (Fig. 3). JRR67 served as a negative control rhAb, while JRR28 and JRR44 were established as strong human and mouse neuron binders in an earlier publication.(Rivas et al., 2017) All three assay methods demonstrated that methods utilizing the acidic elution step reduced or abrogated the ability of JRR44 to bind neurons, whereas neuronal binding by JRR28 was unaffected. It remains unclear how the heavy chain CDR3 charge might influence susceptibility to low pH stress, but as Fig. 4 indicates, antibody gene repertoires do harbor such antibodies and many laboratories interested in their characterization subject rhAbs to low pH stress during elution. This study is limited in that we only tested 2 neuron-binding rhAbs in the experiments. Further evidence of a reproducible effect of acidic elution on neuron binding properties by rhAbs with large negative CDR3 charge is warranted. In addition, we did not test whether 1) other biochemical features of antibody structure and immunogenetics could influence target binding properties, 2) elution with acidic buffers impacts antibody cleavage or degradation, or 3) elution methods influence antigen binding by antibody pools purified from serum or cerebrospinal fluid. Understanding if and how certain biologicals are susceptible to low pH stress is emerging as an important consideration in the development of monoclonal antibodies for therapeutic use in humans. As immunologists interested in identifying antigen targets of antibodies produced by potentially autoreactive B cells, we recommend consideration of low pH stress as a possible confounder in those cases where a rhAb’s antigen target is not identified.
Supplementary Material
Acknowledgements
The authors thank the patients who donated samples for our molecular, cellular and immunogenetics studies of Multiple Sclerosis. We thank all the clinical staff at UTSW for their assistance in sample acquisition and transport. The authors also thank the Whole Brain Microscopy Facility for use of their imaging technology. This work was funded by the NIH to NM (National Institute of Neurological Disorders and Stroke award R01 NS102417) and the Meat Fight Fellowship awarded to NM.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jim.2023.113535.
Data availability
Data will be made available on request.
References
- Blauth K, Soltys J, Matschulat A, Reiter CR, Ritchie A, Baird NL, Bennett JL, Owens GP, 2015. Antibodies produced by clonally expanded plasma cells in multiple sclerosis cerebrospinal fluid cause demyelination of spinal cord explants. Acta Neuropathol. 130 (6), 765–781. Epub 2015/10/30. 10.1007/s00401-015-1500-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabresi PA, 2017. B-cell depletion - a frontier in monoclonal antibodies for multiple sclerosis. N. Engl. J. Med. 376 (3), 280–282. Epub 2016/12/22. 10.1056/NEJMe1614717.28001486. [DOI] [PubMed] [Google Scholar]
- Christley S, Scarborough W, Salinas E, Rounds WH, Toby IT, Fonner JM, Levin MK, Kim M, Mock SA, Jordan C, Ostmeyer J, Buntzman A, Rubelt F, Davila ML, Monson NL, Scheuermann RH, Cowell LG, 2018. VDJServer: A cloud-based analysis portal and data commons for immune repertoire sequences and rearrangements. Front. Immunol. 9, 976. Epub 2018/06/06. 10.3389/fimmu.2018.00976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djoumerska-Alexieva IK, Dimitrov JD, Nacheva J, Kaveri SV, Vassilev TL, 2009. Protein destabilizing agents induce polyreactivity and enhanced immunomodulatory activity in IVIg preparations. Autoimmunity. 42 (4), 365–367. Epub 2009/10/09. 10.1080/08916930902832181.19811303. [DOI] [PubMed] [Google Scholar]
- Friese MA, Schattling B, Fugger L, 2014. Mechanisms of neurodegeneration and axonal dysfunction in multiple sclerosis. Nat. Rev. Neurol. 10 (4), 225–238. 10.1038/nrneurol.2014.37 [DOI] [PubMed] [Google Scholar]
- Gagnon P, Nian R, Leong D, Hoi A, 2015. Transient conformational modification of immunoglobulin G during purification by protein a affinity chromatography. J. Chromatogr. A 1395, 136–142. Epub 2015/04/18. 10.1016/j.chroma.2015.03.080.25882588. [DOI] [PubMed] [Google Scholar]
- Gupta NT, Vander Heiden JA, Uduman M, Gadala-Maria D, Yaari G, Kleinstein SH, 2015. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics 31 (20), 3356–3358. Epub 2015/06/13. 10.1093/bioinformatics/btv359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser SL, Chan JR, Oksenberg JR, 2013. Multiple sclerosis: prospects and promise. Ann. Neurol. 74 (3), 317–327. 10.1002/ana.24009 [DOI] [PubMed] [Google Scholar]
- Imamura H, 2019. pH-shift stress on antibodies. Methods Enzymol. 329–345. [DOI] [PubMed] [Google Scholar]
- Latypov RF, Hogan S, Lau H, Gadgil H, Liu D, 2012. Elucidation of acid-induced unfolding and aggregation of human immunoglobulin IgG1 and IgG2 Fc. J. Biol. Chem. 287 (2), 1381–1396. Epub 2011/11/16. 10.1074/jbc.M111.297697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligocki AJ, Rivas JR, Rounds WH, Guzman AA, Li M, Spadaro M, Lahey L, Chen D, Henson PM, Graves D, Greenberg BM, Frohman EM, Ward ES, Robinson W, Meinl E, White CL 3rd, Stowe AM, Monson NL, 2015. A distinct class of antibodies may be an indicator of gray matter autoimmunity in early and established RRMS patients. ASN Neuro. 10.1177/1759091415609613 pii: 1759091415609613. Print 2015 Sep-Oct.; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez E, Scott NE, Wines BD, Hogarth PM, Wheatley AK, Kent SJ, Chung AW, 2019. Low pH exposure during immunoglobulin G purification methods results in aggregates that avidly bind Fcgamma receptors: implications for measuring Fc dependent antibody functions. Front. Immunol. 10, 2415. Epub 2019/11/05. 10.3389/fimmu.2019.02415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lublin FD, Reingold SC, Cohen JA, Cutter GR, Sorensen PS, Thompson AJ, Wolinsky JS, Balcer LJ, Banwell B, Barkhof F, Bebo B Jr., Calabresi PA, Clanet M, Comi G, Fox RJ, Freedman MS, Goodman AD, Inglese M, Kappos L, Kieseier BC, Lincoln JA, Lubetzki C, Miller AE, Montalban X, O’Connor PW, Petkau J, Pozzilli C, Rudick RA, Sormani MP, Stuve O, Waubant E, Polman CH, 2014. Defining the clinical course of multiple sclerosis: the 2013 revisions. Neurology. 83 (3), 278–286. 10.1212/WNL.0000000000000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzer AR, Perraud X, Halley J, O’Hara J, Bracewell DG, 2015. Protein A chromatography increases monoclonal antibody aggregation rate during subsequent low pH virus inactivation hold. J. Chromatogr. A 1415, 83–90. Epub 2015/09/09. 10.1016/j.chroma.2015.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon MJ, O’Kennedy R, 2000. Polyreactivity as an acquired artefact, rather than a physiologic property, of antibodies: evidence that monoreactive antibodies may gain the ability to bind to multiple antigens after exposure to low pH. J. Immunol. Methods 241 (1–2), 1–10. Epub 2000/08/01. 10.1016/s0022-1759(00)00196-4.10915844. [DOI] [PubMed] [Google Scholar]
- Ortega SB, Torres VO, Latchney SE, Whoolery CW, Noorbhai IZ, Poinsatte K, Selvaraj UM, Benson MA, Meeuwissen AJM, Plautz EJ, Kong X, Ramirez DM, Ajay AD, Meeks JP, Goldberg MP, Monson NL, Eisch AJ, Stowe AM, 2020. B cells migrate into remote brain areas and support neurogenesis and functional recovery after focal stroke in mice. Proc. Natl. Acad. Sci. U. S. A. 117 (9), 4983–4993. Epub 2020/02/14. 10.1073/pnas.1913292117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens GP, Bennett JL, Lassmann H, O’Connor KC, Ritchie AM, Shearer A, Lam C, Yu X, Birlea M, DuPree C, Williamson RA, Hafler DA, Burgoon MP, Gilden D, 2009. Antibodies produced by clonally expanded plasma cells in multiple sclerosis cerebrospinal fluid. Ann. Neurol. 65 (6), 639–649. Epub 2009/06/27. 10.1002/ana.21641.19557869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas JR, Ireland SJ, Chkheidze R, Rounds WH, Lim J, Johnson J, Ramirez DM, Ligocki AJ, Chen D, Guzman AA, Woodhall M, Wilson PC, Meffre E, White C 3rd, Greenberg BM, Waters P, Cowell LG, Stowe AM, Monson NL, 2017. Peripheral VH4+ plasmablasts demonstrate autoreactive B cell expansion toward brain antigens in early multiple sclerosis patients. Acta Neuropathol. 133 (1), 43–60. 10.1007/s00401-016-1627-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothlisberger D, Honegger A, Pluckthun A, 2005. Domain interactions in the Fab fragment: a comparative evaluation of the single-chain Fv and Fab format engineered with variable domains of different stability. J. Mol. Biol. 347 (4), 773–789. Epub 2005/03/17. 10.1016/j.jmb.2005.01.053.15769469. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9 (7), 676–682. Epub 2012/06/30. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman L, 2014. Immunology of relapse and remission in multiple sclerosis. Annu. Rev. Immunol. 32, 257–281. 10.1146/annurev-immunol-032713-120227 [DOI] [PubMed] [Google Scholar]
- Ujas TA, Torres VO, Stowe AM, 2023. Co-culturing immune cells and mouse-derived mixed cortical cultures with oxygen-glucose deprivation to in vitro simulate neuroinflammatory interactions after stroke. Methods Mol. Biol. 2616, 251–260. Epub 2023/01/31. 10.1007/978-1-0716-2926-0_19.36715940. [DOI] [PubMed] [Google Scholar]
- von Budingen HC, Harrer MD, Kuenzle S, Meier M, Goebels N, 2008. Clonally expanded plasma cells in the cerebrospinal fluid of MS patients produce myelin-specific antibodies. Eur. J. Immunol. 38 (7), 2014–2023. Epub 2008/06/04. 10.1002/eji.200737784.18521957. [DOI] [PubMed] [Google Scholar]
- Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC, 2003. Predominant autoantibody production by early human B cell precursors. Science. 301 (5638), 1374–1377 [DOI] [PubMed] [Google Scholar]
- Willis SN, Stathopoulos P, Chastre A, Compton SD, Hafler DA, O’Connor KC, 2015. Investigating the Antigen Specificity of Multiple Sclerosis Central Nervous System-Derived Immunoglobulins. Front. Immunol. 6, 600. Epub 2015/12/10. 10.3389/fimmu.2015.00600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Gilden D, Schambers L, Barmina O, Burgoon M, Bennett J, Owens G, 2011. Peptide reactivity between multiple sclerosis (MS) CSF IgG and recombinant antibodies generated from clonally expanded plasma cells in MS CSF. J. Neuroimmunol. 233 (1–2), 192–203. Epub 2010/12/24. 10.1016/j.jneuroim.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Da RR, Guo W, Ren HM, Hilgenberg LG, Sobel RA, Tourtellotte WW, Smith MA, Olek M, Gupta S, Robertson RT, Nagra R, Van Den Noort S, Qin Y, 2005. Axon reactive B cells clonally expanded in the cerebrospinal fluid of patients with multiple sclerosis. J. Clin. Immunol. 25 (3), 254–264. Epub 2005/06/28. 10.1007/s10875-005-4083-5.15981091. [DOI] [PubMed] [Google Scholar]
- Zhou L, Zhang J, DiGiammarino E, Kavishwar A, Yan B, Chumsae C, Ihnat PM, Powers D, Harlan J, Stine WB, 2018. PULSE SPR: A high throughput method to evaluate the domain stability of antibodies. Anal. Chem. 90 (20), 12221–12229. Epub 2018/09/14. 10.1021/acs.analchem.8b03452.30209948. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.


