Abstract
Surface Plasmon Resonance (SPR) technology, as a powerful analytical tool, plays a crucial role in the preparation, performance evaluation, and biomedical applications of nanoparticles due to its real-time, label-free, and highly sensitive detection capabilities. In the nanoparticle preparation process, SPR technology can monitor synthesis reactions and surface modifications in real-time, optimizing preparation techniques and conditions. SPR enables precise measurement of interactions between nanoparticles and biomolecules, including binding affinities and kinetic parameters, thereby assessing nanoparticle performance. In biomedical applications, SPR technology is extensively used in the study of drug delivery systems, biomarker detection for disease diagnosis, and nanoparticle-biomolecule interactions. This paper reviews the latest advancements in SPR technology for nanoparticle preparation, performance evaluation, and biomedical applications, discussing its advantages and challenges in biomedical applications, and forecasting future development directions.
Keywords: SPR, nanoparticles, biomedical applications, nanomaterials, characterization
Introduction
Nanomedicine represents the utilization of nanotechnology in the medical field, with the goal of innovating diagnostic, therapeutic, and preventive approaches to enhance treatment efficacy and precision. Nanomedicine research focuses on the development, properties, and medical applications of nanoparticle materials. Several therapeutics for cancer, pain and infectious diseases have been successfully developed based on nanoparticles (particles with size range from 1 nm to 1000 nm),1 including metal nanoparticles, lipid-based nanoparticles, as well as polymeric nanoparticles.1–4 Nanoparticles have been frequently studied for application in various medical setting, including drug delivery,5 targeted therapy,6,7 imaging enhancement,8 and early detection of disease.9 To construct effective nanoparticle systems for biomedical applications, the nanoparticle surface characteristics and the interactions with biological systems need to be thoroughly characterized and tested. To analyze interactions between biomolecules, various methods have been developed. Electrophoretic mobility shift assays (EMSA) and co-immunoprecipitation (CoIP), however, are complex and prone to errors.10 Techniques like enzyme-linked immunosorbent assays (ELISA), fluorescence resonance energy transfer (FRET), and microscale thermophoresis (MST) require labeling, which may affect molecular activity.11,12 Although isothermal titration calorimetry (ITC) can be used for label-free evaluation of binding affinity, it requires large sample volumes and is unsuitable for monitoring interactions in small samples. SPR is an optical detection method that measures changes in the refractive index near the surface of a material film coated on a glass substrate. These changes indicate molecular interactions occurring at the surface. As a label-free optical technology, SPR detects interactions based on changes in refractive index, reducing potential interference and allowing accurate assessment of binding properties.13 SPR technology can provide detailed characterization of the surface functionalization of nanoparticles and allow observation of interactions between nanoparticles and biomolecules for studying the behavior and mechanisms of nanoparticles within biological systems. SPR can detect extremely low concentrations of biomolecules, providing precise data and reliable results when investigating interactions between nanoparticles and target molecules (such as proteins, DNA, RNA, etc). Therefore, SPR technology plays an indispensable role in the research of biomedical nanoparticles.
SPR, serving as a technique for the rapid screening of molecular interactions, holds tremendous potential in aiding the research and development of nanoparticles as drug delivery systems and nanomedical diagnostic devices. In recent years, SPR technology has rapidly advanced, with a focus on the development and construction of novel biosensor chips.14,15 Customized biosensors have been created for qualitative and quantitative measurements in both artificial and real samples, yielding results consistent with the gold standard method of liquid chromatography-tandem mass spectrometry (LC-MS/MS).16,17 The construction of high-throughput microplate biosensors based on 3D nanocup array structures enables high-throughput screening, reducing costs and increasing speed. The integration of nanomaterial-enhanced particles amplifies detection signals, significantly improving sensitivity and making the process more convenient, sensitive, and efficient than traditional ELISA.18 Additionally, incorporating two-dimensional materials such as graphene, transition metal dichalcogenides (TMDCs), MXene, black phosphorus (BP), metal-organic frameworks (MOFs), and antimonene into sensors can substantially enhance their sensitivity and detection performance.19–21 The use of optical fiber sensing probes with PAN nanofiber membranes and gold nanofilm composite sensitive membranes allows for high sensitivity and selectivity in detection.22 Furthermore, the combination of alternating current (AC) effects and surface plasmon resonance (SPR) in rapid, high-sensitivity biosensors integrates AC electro-osmosis (ACEO) and dielectrophoresis (DEP) phenomena within the SPR biomarker sensing region, improving the efficiency of target analyte collection.23
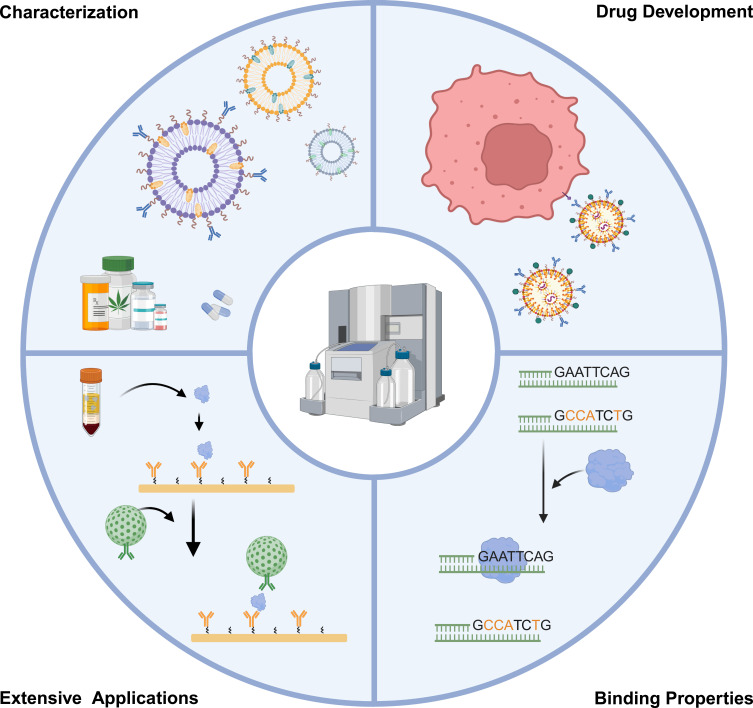
In this review, we provide an overview of the SPR technology applied in the biomedical field, including nanoparticle characterization, drug development, binding properties, as well as extensive application, as shown in Figure 1. The aim is to promote a more precise and extensive application of SPR technology, thereby advancing research, therapeutics, and diagnostic devices based on nanoparticles.
Figure 1.
SPR technology applied in the biomedical field, including nanoparticle characterization, drug development, binding properties and extensive application.
SPR Equipment and Working Principles
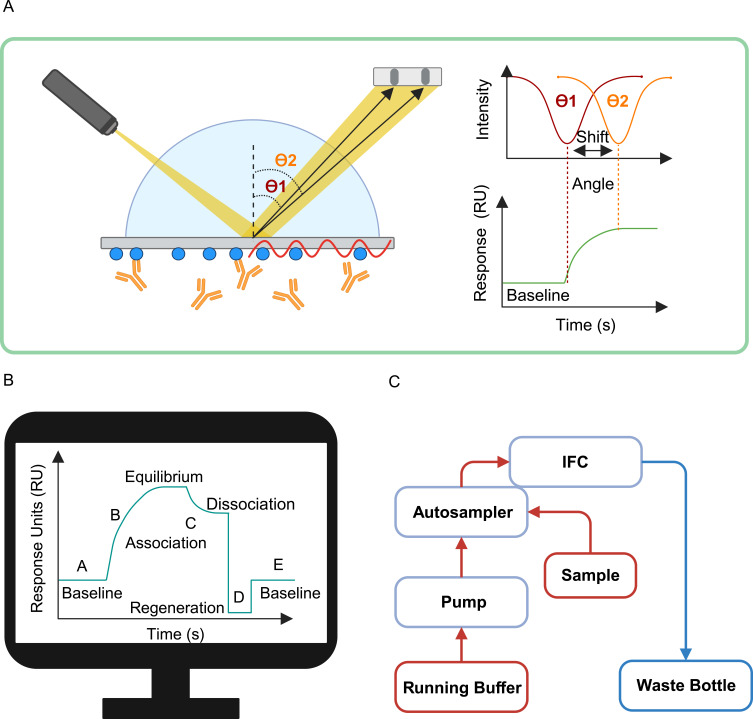
SPR devices consist of three primary components: an optical detection system, sensor chips, and a microfluidic system. Figure 2 shows the main components of SPR instrument. The optical detection system is utilized to detect changes in refractive index near the surface of sensor chips. During molecular interactions, binding between molecules alters the refractive index, causing a proportional shift in incident light angle relative to the mass of bound molecules, enabling sensitive detection of molecular interactions. Sensor chips interact with samples through surface-modified functional groups and detect resonance changes using an external light source. Interaction between sensor chips and samples occurs through methods such as covalent attachment, high-affinity capture, and hydrophobic adsorption, tailored according to specific application needs. The microfluidic system precisely delivers buffer solutions and samples to ensure their uniform distribution on the sensor chip, thereby enhancing detection sensitivity. The refractive index sensing mode is the most commonly used SPR detection mode. It detects the binding or reaction events of biomolecules by monitoring the refractive index changes on the surface of the sensing layer. With the development of SPR technology, various SPR detection modes have been developed to meet specific application needs.24 The spectral interrogation mode analyzes the interactions between biomolecules and the sensing layer by detecting the wavelength or frequency changes of the SPR signal.25,26 The angle scanning mode determines the binding situation of biomolecules to the sensing layer by adjusting the incident light angle to detect the maximum change in the SPR signal.27,28 The polarization interrogation mode studies the directional binding and orientation of biomolecules on the sensing layer by changing the polarization state of the incident light.29–31 Additionally, the time scanning mode monitors the SPR signal changes over time and is used for dynamic studies of the binding and release kinetics of biomolecules.32 These different SPR detection modes have broad applications in biomedical research and applications, providing detailed information on biomolecular interactions, binding kinetics, and the interactions between nanoparticles and biological systems.
Figure 2.
The main components of SPR instrument. (A) Optical module of SPR: The change of refractive index causes the angle shift from θ1 to θ2. (B) Signal collection system: signals collected at different binding stages. (C) Schematic of microfluidic system, arrows indicate the direction of fluid flow through the system.
SPR in Nanoparticle Research for Biomedical Application
SPR technology has become a pivotal tool in the nanoparticle research for biomedical applications. SPR aids in elucidating molecular interactions at the nanoscale, providing deep insights into fundamental biological processes and promoting the development of new therapies. The technology enables real-time, label-free monitoring of molecular interactions, which is crucial for understanding the mechanisms of drug delivery systems. SPR-enhanced sensors have revolutionized biosensing technology by detecting trace amounts of biological substances, thereby supporting rapid and accurate diagnostics. SPR offers unparalleled sensitivity and specificity for the detection of biomolecules, making it indispensable for early disease diagnosis. The SPR technology applied in nanoparticle studies is summarized in Table 1.
Table 1.
Summary of SPR Technology Applied in the Nanoparticle Studies
| Nanoparticles Type | Analyte | Immobilized Agent | Application | References |
|---|---|---|---|---|
| Liposomes | Different composed liposomes | Aβ1-42 fibrils | Properties characterization | [33] |
| Liposomes | PEGs liposome | HSA, ApoE, α2-M, β2-G, Fn | Properties characterization | [34] |
| Liposome | Glucose | Glucose-imprinted liposomes | Drug Development and Screening | [35] |
| Liposome | PEG-Liposome, RGD-Liposome, TH-Liposome | Integrin αvβ3 | Drug Development and Screening | [36] |
| Liposomes | bFGF, bFGFp-liposome, bFGFp-liposome + bFGF | FGFR1 | Drug binding properties | [37] |
| Liposomes | Anti-MUC-1- Temperature-sensitive liposomes | Scrambled MUC-1 | Drug binding properties | [38] |
| Liposomes | Anti-ephrin A2- Liposomes | Ephrin A2 extracellular domain | Drug binding properties | [39] |
| Liposomes | PEGylated liposomes | G-CSF, FVIII, FVIIa | Drug binding properties | [40] |
| Liposome | hCD40L + peptide | CD40 | Drug binding properties | [41] |
| Liposome | Cyclic RGD motifs-liposomes | Melanoma B16 and MCF-7 cells | Mechanism of drug therapy | [42] |
| Liposomes | Cationic liposomes | S. mutans biofilm | Mechanism of drug therapy | [43] |
| Liposomes | BsmAb hMN14x734 | Types of liposomes | Properties characterization | [44] |
| Liposome | Nex-1 | Different liposomes | Drug Development and Screening | [45] |
| Liposome | Etx | Different liposomes | Drug Development and Screening | [46] |
| Liposome | Salmeterol and propranolol | Liposome | Mechanism of drug therapy | [47] |
| Liposome | Sulfated 3-O-octadecyl-(1→6)-α-D- glucopyranan, sulfated alkyl maltoheptaoside | Liposome | Mechanism of drug therapy | [48,49] |
| Liposome | Detergent | Liposome | Mechanism of drug therapy | [50] |
| Liposome | NRG1+ ErbB4-coated liposomes | Biotin Anti-IgG-Fc | Extension Applications | [51] |
| Liposome | sfGFP-ANXV, ANXV, sfGFP | Phospholipid liposomes | Extension Applications | [52] |
| Liposome | OptoPB, V416 variants | POPC liposomes | Extension Applications | [53] |
| Liposomes and lipid nanoparticle | Anti-CD163 antibody- Liposomes, Anti-CD163 antibody lipid nanoparticle | CD163 | Drug binding properties | [54,55] |
| Lipid nanoparticle | Anti-HB-EGF-modified LNP-siRNA | rhHB-EGF | Drug binding properties | [56] |
| Lipid Nanoparticle | HA- Lipid Nanoparticle | CD44-Fc chimera | Properties characterization | [57] |
| Liposome membranes | Cry4Ba-DIII truncate Cry4Ba full-length toxin |
Liposome membranes | Drug binding properties | [58] |
| Nanoliposomes | Curcumin-nanoliposomes | Aβ1-42 monomers or fibrils | Properties characterization | [59] |
| SMA liposomes | Potential P-gp inhibitor | SMALPs-MCF-7, SMALPs MCF-7/ADR | Extension Applications | [60] |
| Lipid Salipro nanoparticles | Carbenoxolone, spironolactone, benzoylbenzoyl-ATP | His/Biotin-Salipro-empty, His/Biotin-Salipro-mPANX1 | Extension Applications | [61] |
| Exosomes | Rituximab obinutuzumab | CD20 exosome | Extension Applications | [62] |
| Exosomes | InP/ZnS-AB + EBC-derived EVs | Goat anti-Mouse IgG1 | Extension Applications | [63] |
| Engineered exosome | PD1-exosome | PDL1 | Drug Development and Screening | [64] |
| Polycationic Albumin Nanoparticles | HAS, lipopolysaccharides | HSA and PEG (2000)18-cHSA | Properties characterization | [65] |
| Molecularly imprinted nanoparticles | Andarine, Ligandrol, RAD-140 | Different SARMs nanoMIPs. | Extension Applications | [66] |
| PAMAM-based nanoparticles | Folic acid- PAMAM-based nanoparticles | Folic Binding Protein | Properties characterization | [67] |
| MIP nanoparticles | Trypsin-specific MIP nanoparticles | Trypsin | Properties characterization | [68] |
| Spike trimer-ferritin fusion recombinant protein nanoparticle | Sera of mice | S-2P and RBD | Drug Development and Screening | [69] |
| Kappa spike and RBD mRNA-LNP | SARS-CoV-2 BA.1, BA.2 RBD protein | K-RBD chAbs | Drug Development and Screening | [70] |
| N-S1 protein double-layered nanoparticle | Soluble S1 protein, N-S1 PNp | ACE2-His | Drug Development and Screening | [71] |
| Mosaic receptor-binding domain (RBD) nanoparticle | Mosaic_RBD-NP | hACE2 | Drug Development and Screening | [72] |
| Polypeptide-based nanoparticles | Nanoparticles Self-assembling polymers | Bacterial cells | Mechanism of drug therapy | [73] |
| Adamantane derivatives self-assembled nanoparticles | SNPs | Biotinylated melittin | Drug Development and Screening | [74] |
| Chitosan nanoparticles | Transferrin-modified chitosan nanoparticles | His-tagged Transferrin receptor | Drug binding properties | [75] |
| Protein nanoparticle | Albumin peptides, Bevacizumab, Rituximab, and Trastuzumab | Rituximab, albumin peptides | Drug binding properties | [76,77] |
| Gd-chelated polysiloxane nanoparticles | AGuIX NPs, AGuIX@A12 VHH, AGuIX@A4 VHH | PD-L1 and CD47 | Extension Applications | [78] |
| rPAA-Chol nanoparticles | rPAA-Chol nanoparticles | Liposomes | Mechanism of drug therapy | [79] |
| Nanobody | aSA3 and aRBD-2 | RBD | Drug binding properties | [80] |
| PFC nanoparticles | NBD-Linker | PFC nanoparticle | Mechanism of drug therapy | [81] |
| Biotin-FNPs | PSA + Ab2-SA-biotin-FNP | Ab1 | Extension Applications | [82] |
| HA-PEI-coated ECH-Zn (PPzn) | ECH-Zn | STAT2, MDM2, STAT2600−750, STAT2Δ600−750 | Drug binding properties | [83] |
| Silica nanoparticles | Biotinylated- silica nanoparticles | Streptavidin | Properties characterization | [84] |
| AuNPs, silica NPs | SAgs +anti-SEG Abs coupled nanoparticles | Purified antibodies | Extension Applications | [85] |
Characterization of Nanoparticles Properties
Size of Nanoparticles
The diameter of nanoparticle carriers plays a pivotal role in pharmacokinetics, as their size determines the potential to traverse biological barriers within the body. SPR technology can detect the size of nanoparticles by measuring the affinity between substances and optimize their diameter. For example, biotin and streptavidin (with KD values of 1×10−15 M and 1×10−13 M, respectively), as well as antigen and antibody interactions (with KD values of 1×10−12 M and 1×10−9 M), are recognized for their high affinity. By labeling silicon nanoparticles of different diameters (50, 100, and 200 nm) with biotin and flowing them over a chip surface coated with streptavidin using a single-cycle kinetics method, then comparing them with positive KD values, nanoparticles of sizes more conducive to intermolecular interactions can be screened. This methodology facilitates the selection of nanoparticle sizes that optimize biological interactions, thereby enhancing the efficacy of nanoparticle-based drug delivery systems.84
Composition of Nanoparticles
SPR can also screen for the optimal components of liposomal nanoparticles based on affinity comparison. In the study of amyloid-beta (Aβ) peptides as potential targets for Alzheimer’s disease (AD) treatment,86 by immobilizing Aβ1-42 fibrils and comparing the specific binding of different component liposomes to amyloid-beta (Aβ), it was found that liposomes embedded with 5% or 20% phosphatidic acid (PA) and liposomes with 5% or 20% cardiolipin (CL) have nanomolar-level affinity with Aβ1-42 fibrils, compared to pure liposomes. This confirms that PA and CL could be the best candidate headgroup components for Aβ1-42. This demonstrates the utility of SPR in identifying and optimizing nanoparticle formulations for targeted therapeutic applications, highlighting its potential in advancing treatment strategies for diseases like AD.33
Surface Modification of Nanoparticles
SPR technology can also be used to detect chemical modifications or functionalization on the surface of nanoparticles. It enables the screening for the presence,65 quantity,34,67 molecular weight,57 and location of surface modifications,44 as well as characterizing the structure of the modifiers.59 For example, to obtain the most effective polyethylene glycolated (PEGylated) liposomes for monoclonal bispecific antibodies (BsmAb, anti-CEA (carcino-embryonic antigen) x anti-DTPA-In (diethylenetriaminepentaacetic acid-indium)), Rauscher et al conducted optimizations. They optimized the liposomal nanoparticles by adjusting the length and concentration of PEG chains, as well as the position of hapten modifications. These differently modified liposomes were immobilized on an L1 chip, and diluted BsmAb was injected from low to high concentrations in a multi-cycle kinetic mode, eventually fitting the KD values with different modified liposomes. The results indicated that when the DTPA-In hapten was coupled to the end of the PEG chain (DSPE-PEG-DTPA), it exhibited the best affinity (KD = 6.3 nM). This demonstrates SPR’s capability in fine-tuning the functionalization of nanoparticle surfaces to enhance their interaction with specific targets, providing a crucial tool for optimizing nanomedicine formulations.44
Stability of Nanoparticles
SPR technology can be used to assess the stability and consistency of nanoparticles under different treatment conditions. Biomaterials often utilize lyophilization and sterilization as the most common methods of storage. To evaluate the impact of lyophilization and sterilization on the recognition characteristics and stability of Nanoparticles (NPs), SPR assessed the interaction between molecularly imprinted polymer nanoparticles (MIP NPs) and trypsin before treatment (KD=15.8 nM), and then with MIP NPs after lyophilization (KD=7.1 nM, Chi2 = 1×10−5) and after sterilization (KD = 12.2 nM, Chi2 = 2.3×10−4). The results showed that lyophilization and sterilization treatments did not affect the activity of the MIP NPs, indicating that harsh conditions (lyophilization and sterilization) are suitable for the long-term storage of MIP NPs.68 This demonstrates the utility of SPR in ensuring the viability of nanoparticle-based technologies through storage and handling processes.
Drug Development and Screening
Drug development is a complex scientific process aimed at discovering, designing, testing, and promoting new medications for treating and alleviating symptoms of diseases. Given the rapid transmissibility and significant pathogenicity of SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus 2), there is an urgent need to develop a broad-spectrum medication that can prevent upper respiratory infections. Particularly, the rapid mutation of SARS-CoV-2 challenges the effectiveness of some antibody medications, while vaccines based on nanoparticles show great potential to overcome the limitations of existing vaccine technologies. The global scientific community is engaged in the development of nanoparticle medications against COVID-19 (Coronavirus Disease 2019) constructing stable immune platforms through the rapid screening process of SPR technology,69,87 and developing various nanoparticle immunotherapeutics aimed at eliciting stronger immune responses in the body, prompting the production of a large number of neutralizing antibodies to resist viral invasion.71,72
In the field of cancer treatment, although immune checkpoint therapies have enabled long-term remission and prevented the clinical recurrence of cancer symptoms in some patients, more than half of the patients have failed to effectively respond to this therapy due to immune exhaustion. This study proposes an innovative genetically engineered exosome strategy, which involves engineering the PD1 (Programmed death-1) gene and encapsulating the immune adjuvant imiquimod (PD1-Imi Exo). Through SPR experiments, PD1 Exo demonstrated significant high affinity with PDL1 (Programmed death-ligand 1) protein and was found to effectively reverse T-cell exhaustion, providing new insights and strategies for preventing postoperative tumor recurrence or metastasis.64
Drug screening is a critical process for assessing the efficacy and safety of potential drug candidates in treating specific diseases, essential for evaluating drug synthesis to ensure the most promising candidates can further be developed into effective medications. Nanoparticles are often used as drug delivery systems to improve the pharmacokinetics and biodistribution of drugs and reduce their toxicity.88 In the pursuit of efficient nanoparticle drug discovery, SPR technology has demonstrated its irreplaceable value. For instance, to rapidly discover effective inhibitors of melittin, researchers utilized a system of self-assembling nanoparticles (SNPs) formed by cyclodextrin polymers and adamantane derivatives. They immobilized biotinylated melittin on a chip surface containing avidin and rapidly screened eight self-assembling nanoparticles from a candidate drug library that could bind and neutralize the toxicity of melittin using SPR technology.74 This step not only expedited the discovery process of inhibitors but also enhanced the precision of the screening process. Furthermore, the application of SPR technology extends to developing clinically useful, long-term implantable, self-regulating insulin delivery systems. By incorporating amino acid derivatives (dihexadecyl-glutamate-glutamine (DHD-glu-gln), dihexadecyl-glutamate-asparagine (DHD-glu-asn), dihexadecyl-glutamate-glutamic acid (DHD-glu-glu)) into liposomes, molecules sensitive to glucose were formed. Facing the challenge of rapidly screening the most effective formulations, the authors used SPR technology to immobilize the amino acid derivatives and their liposomes on an L1 chip. They collected and compared signal changes for each sample with a glucose solution. This method allowed researchers to qualitatively assess the binding affinity of amino acid derivative liposomes as glucose-sensitive molecules, providing a scientific basis for the rapid development of insulin delivery systems.35
Moreover, SPR technology can be used for the screening of interaction conditions between two molecules, allowing the evaluation of nanoparticles’ affinity with target molecules under varying pH and calcium ion concentrations,36,45,46,89 as well as other environmental conditions. This enables rapid determination of which conditions are more conducive to interactions between substances. These applications of SPR contribute to a deeper understanding of the interactions between drugs and biomolecules and the development of new therapeutic strategies.
In summary, SPR technology holds significant promise in drug development and screening due to its real-time, label-free analysis of molecular interactions.18,60 SPR facilitates the rapid high-throughput screening of extensive libraries of drug candidates by immobilizing target molecules on the sensor surface and measuring binding interactions simultaneously, thus accelerating early-stage drug discovery.18,90 This technology also provides detailed kinetic information for understanding binding mechanisms and selecting candidates with optimal characteristics.91 The real-time monitoring capability of SPR allows researchers to observe dynamic processes and transient interactions that might be missed by endpoint assays, providing valuable insights into the temporal aspects of drug binding and efficacy.92,93 Moreover, SPR aids in optimizing lead compounds by analyzing the binding interactions of various analogs and derivatives, helping identify structural features that enhance binding affinity and specificity.94 It also supports biophysical characterization, such as binding stoichiometry and conformational changes, offering insights into the mechanism of action and predicting potential off-target effects.95,96 SPR technology can be integrated with other analytical techniques, such as mass spectrometry or nuclear magnetic resonance (NMR), to provide complementary data for a comprehensive understanding of drug interactions and the development of multi-target therapies.97,98 In personalized medicine, SPR can screen patient-specific biomolecules to identify the most effective therapeutic agents, tailoring treatments based on individual molecular profiles and contributing to more effective healthcare solutions.99
Exploring Drug Binding Properties
SPR technology is also utilized in the study of drug binding characteristics of nanoparticle medicines, playing a crucial role in revealing drug targeting mechanisms, identifying drug binding sites, exploring drug binding properties, and analyzing epitope competition between drugs. This capability not only facilitates the discovery and development of new drugs but also provides precise molecular insights for drug design.
Targeting
NPs as drug delivery systems aim to transport drugs to desired biological targets, making the targeting ability of NPs crucial for accurate drug delivery. SPR technology has been widely applied to characterize the targeting functionality of NPs. Typically, this involves coupling target molecules to the sensor chip, then using targeted molecules or drugs with targeting modifications,56,100 ultimately predicting the in vivo targeting of the drug based on the strength of affinity. The active targeting of NPs relies on targeting molecules modified on the NPs surface. These targeting molecules are often ligands for receptors at target sites within the body,37,75,101,102 as well as antigens or antibodies in antigen-antibody pairs,38,39,54,55 utilizing their specific binding to achieve targeted in vivo delivery of nanoparticle drugs.
Binding Sites
Researchers have utilized SPR technology to identify specific binding sequences of blood proteins that interact with polyethylene glycol-modified liposomes (PEGLip). By immobilizing purified proteins on a chip and conducting SPR analysis with PEGLip, it was discovered that proteins binding to PEGLip share a specific amino acid sequence of 8 residues (S/T-X-L/V–I/Q/S-S/Q/Q/T/Q-X-X-E). Peptides containing this sequence were synthesized, and their binding affinity to PEGLip (KD=2.3 nM) was found to be very close to that of the full-length proteins. This confirmed that short sequences containing specific amino acids are sufficient for binding to PEGLip.40
To further explore drug characteristics and binding mechanisms, Nevala et al developed an antibody-targeted nanoparticle chemotherapy delivery strategy. This strategy involves non-covalently attaching Rituximab (targeting CD20) to an albumin-bound paclitaxel (ABX) scaffold, aiming for targeted recognition of tumors. To determine the precise binding sites and regions between Rituximab and the ABX albumin scaffold, the research team utilized SPR. A library of albumin peptides containing 18 amino acids was covalently fixed on the chip, identifying peptide 40 that binds to Rituximab, located in the Sudlow II hydrophobic binding domain of albumin.76 Conversely, to explore the binding sites of albumin on different anti-CD20 antibodies, peptide 40 was used as the immobilized ligand, and different concentrations of Bevacizumab peptides were screened through SPR, revealing the CDR H3 loop of the Fab region as the shared binding domain for Bevacizumab, Rituximab, and Trastuzumab.77 These findings not only determined the binding sites of monoclonal antibodies on albumin but also identified the binding regions of albumin on various monoclonal antibodies, providing solid theoretical support for precise drug design.
SPR technology was utilized to elucidate the mechanism through which PPZn nanoparticles (HA-PEI (hyaluronic acid/poly(ethylenimine))-coated ECH-Zn (echinacoside-zinc)) exert their antiglycation effect by regulating RAGE (Advanced glycosylation end product-specific receptor) via MDM2 (mouse double minute 2) and STAT2 (Signal transducer and activator of transcription 2).83 SPR experiments revealed that ECH-Zn binds directly to STAT2 in a concentration-dependent manner, with no significant affinity observed between ECH-Zn and MDM2. To further validate the binding region of ECH-Zn with STAT2, truncated protein experiments involving STAT2600−75 and STAT2Δ600−750 were conducted. The results indicated that ECH-Zn interacts with STAT2600−750, but loses activity towards STAT2Δ600−750. This suggests that ECH-Zn achieves its antiglycation effect by binding to STAT2600−750, thereby blocking the interaction between STAT2 and MDM2 and reducing the activation of RAGE.
In research exploring insecticidal mechanisms, SPR technology was employed to detect the interaction between Cry toxins and lipid nanoparticles, thereby identifying the role of protein functional domains.58 Researchers focused on the DIII truncated protein of the Cry toxin and found through SPR that both the Cry4Ba-DIII truncation and its full-length protein could tightly bind to liposomes immobilized on an L1 chip. To further investigate their mechanism of action, a protease K digestion SPR experiment was conducted. During the dissociation phase, under the action of protease K digestion, the binding strength of Cry4Ba-DIII and the full-length toxin was reduced compared to bovine serum albumin (BSA), but still remained at a higher level (Figure 3). This indicates that parts of these proteins indeed embed into the lipid layer interior, thus avoiding digestion by protease K. This experiment not only confirmed the crucial role of the Cry4Ba-DIII truncation in cell membrane insertion and target recognition but also showcased the application value of SPR technology in analyzing the interaction between lipid nanoparticles and proteins and determining protein functional domains. It provides profound insights into understanding complex biological interactions.
Figure 3.
Real-time Sensorgram obtained from a proteinase K protection assay with Sample (red) and BSA (green). The injection of proteinase K into individual membrane-associated proteins is indicated by arrows. Horizontal arrows show the resonance unit (RU) values at the start and end of the proteinase K injection phase.
Binding Epitopes
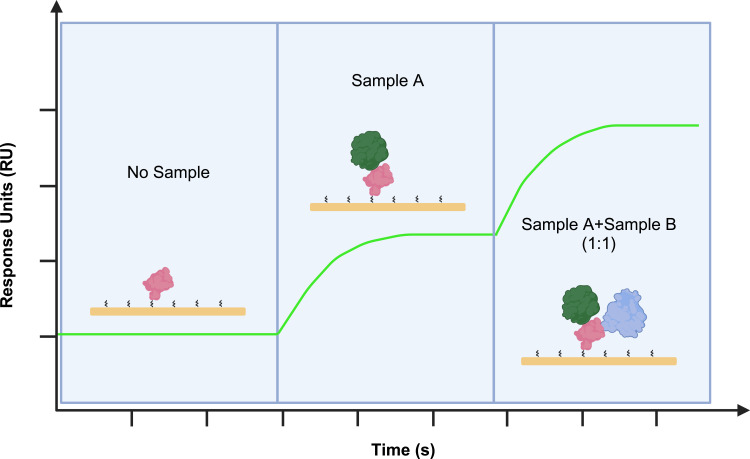
Beyond analyzing interactions between two molecules, the flexibility of SPR technology extends to detecting interactions among three molecules, which is crucial for unveiling complex binding mechanisms. In research exploring interactions among three molecules, SPR technology can be used to identify different binding epitopes (Figure 4).
Figure 4.
Fixation of target protein on a CM5 chip via SPR to observe the binding of Sample A and Sample B to target protein. Initially, Sample A is injected for 120 seconds, followed by the injection of a 1:1 mixture of Sample A and Sample B for 120 seconds to observe to different binding epitopes.
In the development of COVID-19 antibody drugs, SPR technology has been successfully applied to ascertain whether bispecific antibodies can specifically bind to different epitopes of the receptor-binding domain (RBD) of SARS-CoV-2 (SARS2).80 Researchers immobilized SARS2 RBD on a CM5 chip, first introducing the aSA3 antibody to observe its binding with RBD. Upon introducing a mixture of aSA3 and aRBD-2, a significant increase in signal was detected, clearly indicating that the two antibodies bind to different regions of the RBD rather than competing for the same binding site. This not only confirms the bispecific binding characteristics of aSA3 and aRBD-2 on nanoparticles but also highlights SPR as a powerful in vitro tool for analyzing the binding epitopes of antibody-target protein interactions, underscoring its application potential in antibody research and development.
Surface Competition
In the study of competitive interactions, SPR also offers significant advantages. Taking CD40L (Cluster of Differentiation 40-Ligand) as an example, this protein is a key target for the treatment of autoimmune diseases. The interaction between CD40L and CD40 has been extensively studied with the aim of developing antibodies or other molecules that can interrupt this interaction for therapeutic purposes. To construct peptide ligands as blockers, Ding et al utilized the crystal structure information of CD40L and molecular docking techniques to design six peptide ligands with high specificity for binding to CD40L. By conducting competition binding experiments using SPR technology, with CD40 immobilized on the chip and mixing hCD40L with various peptide ligands, they measured the competitive binding situation of hCD40L with these peptide ligands, identifying peptide ligands that could efficiently block the interaction between CD40 and CD40L.41 Subsequently, using the identified ligands, liposomes loaded with the cell-inhibitory drug methotrexate (MTX) were modified, which was found to significantly improve clinical scores. This method not only identified peptide ligands with the best competitive effect but also highlighted how SPR provides crucial experimental evidence for drug design and optimization targeting specific disease markers.
SPR technology offers significant promise for studying drug binding properties due to its capacity for real-time, label-free analysis of molecular interactions.103,104 This technology allows for precise measurement of binding kinetics, including association and dissociation rates, and affinity constants, providing detailed insights into the binding mechanisms of potential drugs.62 SPR’s high sensitivity and versatility enable the study of a diverse range of interactions, such as protein-protein, protein-DNA, and protein-small molecule interactions.77,105 This makes it an invaluable tool for optimizing drug candidates by identifying those with optimal binding characteristics, understanding their biophysical properties, and observing dynamic and transient interactions that are often missed by traditional methods.106,107
Investigating the Mechanism of Drug Therapy
The bioavailability of a drug significantly depends on its solubility in cell membranes, and predicting or quantifying this characteristic is crucial for precise drug design. In the study of nanoparticle drug release processes, the application of SPR technology can directly immobilize biological cells on the chip surface to simulate an in vitro environment for exploring the interactions between nanoparticle drugs and cell membranes, as well as drug release mechanisms. For instance, Guo et al covalently attached cell surface proteins from B16 and MCF-7 tumor cells to the sensor chip, successfully simulating the direct interaction between nanoparticle drugs and cell membranes. By injecting liposomes modified with different structures of RGD peptides, researchers were able to accurately assess the interaction strength between various liposomes and tumor cell membranes.42
Furthermore, the stable binding was observed between nanoparticles and bacterial cell membranes by directly immobilizing bacterial cells on a gold sensor chip. They confirmed that nanoparticles have a stronger affinity to adhere to bacterial surfaces than traditional antibiotics and are difficult to wash off. This allows them to penetrate membrane structures and cause leakage of cell contents, thereby exerting their broad-spectrum antibacterial effects.73 Compared to traditional liposomes, cationic liposomes can engage in stronger interactions with bacterial membranes,43 and SPR detection results also demonstrated that cationic liposomes capable of generating electrostatic interactions could be developed and utilized as a potential drug delivery system for biomembrane.
SPR has also been used to study the membrane binding behavior of drugs. By coupling DOPC liposomes to the chip surface, multicycle kinetic analysis was conducted on nearly 100 drugs to categorize their interactions with lipid bilayers into three types: non-binding, reversible binding, and stable complex formation.108 Using liposomes as model cell membranes not only deepens our understanding of the interaction mechanisms between drugs and cell membranes but also offers a new perspective for evaluating the release rate of drugs from modified nanoparticles,47,79,81 facilitating the further development of drugs targeting specific diseases.48,49 Given the complex behavior of liposomes in vivo, devising testing methods in vitro that can perfectly simulate their release behavior in vivo presents a challenge. An innovative approach was adopted, by immobilizing different liposomes on an L1 chip and flowing different concentrations of detergent over the liposome surface to simulate the environment in vivo. The study found that the nonionic surfactant TX-100 significantly increased the membrane permeability and solubility of PEGylated liposomes.50 The application of SPR technology to assess lipid membrane instability induced by detergents, and consequently drug release, showcases an efficient and time-reducing new pathway for evaluating the properties of liposome membranes.
Extension of SPR in Nanoparticle Researches
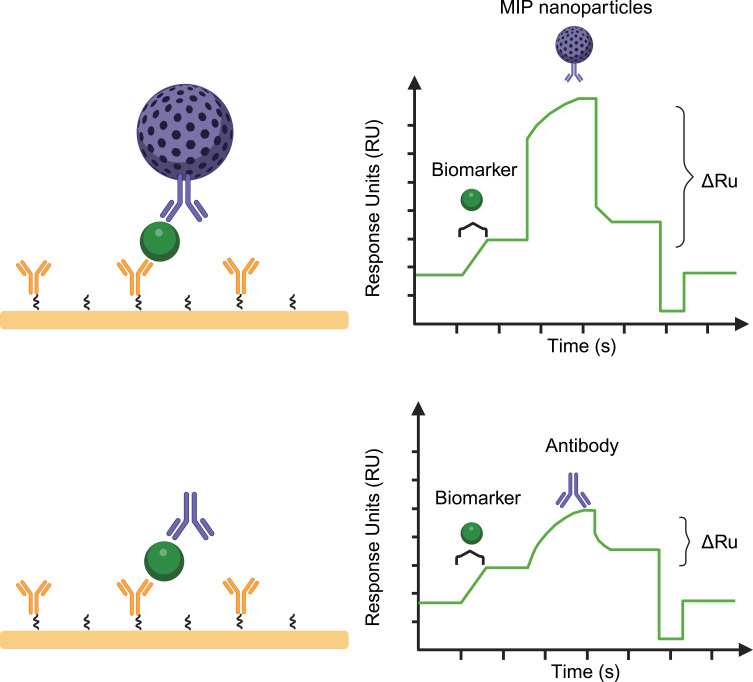
Leveraging the high sensitivity and real-time monitoring capability of SPR, combined with the diverse applications of nanotechnology, provides a powerful detection method and tool for life sciences and diagnostic medicine research. SPR significantly enhances detection sensitivity, making it possible to detect trace biomolecules such as viral sequences, disease markers, and cell abnormalities. The integration of this technology offers an effective means to identify and amplify the signal of target analytes (Figure 5), driving the application of SPR technology in nanoparticle research to reveal more extensive uses.
Figure 5.
Schematic illustration of molecularly imprinted nanoparticles enhanced biomarker detection using an SPR biosensor. Injection of biomarker, followed by injection of molecularly imprinted nanoparticles.
Reliable and sensitive detection of biomarkers is crucial, especially for circulating biomarkers with short half-lives. The development and preparation of SPR combined with nanoparticles have significant advantages in the detection of low-concentration molecules. Reliable and sensitive detection of biomarkers is crucial, especially for circulating biomarkers with short half-lives. To address this challenge, researchers developed a detection platform combining molecularly imprinted nanoparticles with SPR to monitor the expression levels of neuregulin 1 (NRG1) in biological samples. The platform ensures precise binding of Fc-NRG1 by directionally coupling biotinylated anti-IGG-Fc (Hu) to an SA chip. By injecting specifically modified ErbB4 liposomes, the multivalent interactions significantly enhance the affinity signal, achieving high-sensitivity detection of trace amounts of NRG1 in blood samples, with a detection limit as low as 3.5 pM. This represents about a 60-fold improvement over traditional SPR and immunoassay methods.51 Furthermore, the same approach, using nanoparticles with different molecular imprints, can also be applied to detect various Selective Androgen Receptor Modulators (SARMs) in athletes’ bodies,66 as well as trace markers in other biological samples.82,85 Therefore, nanoparticles enhance the SPR signal, enabling high-sensitivity detection and quantitative analysis of trace biomolecules. SPR biosensors based on molecularly imprinted nanoparticles exhibit good accuracy, high specificity, and high repeatability, capable of directly detecting small amounts of target components in samples within a short time. This innovative method not only breaks through the sensitivity limitations of biomarker detection but also provides strong technical support for precision medicine, food testing, environmental monitoring, and more.
Membrane-binding proteins hold a crucial place in drug development, with many existing drugs designed to target these proteins. SPR technology has significant applications in the development of drugs targeting membrane-bound proteins. By monitoring the interactions between membrane proteins and candidate drug molecules in real-time, SPR provides label-free, high-sensitivity detection, aiding researchers in identifying and screening potential drug targets and inhibitors. The overexpression of P-glycoprotein (P-gp) in cancer cells can trigger the phenomenon of multidrug resistance (MDR), and the combined use of anticancer drugs with P-gp inhibitors is considered an effective strategy to reverse MDR. However, the challenge of purifying and immobilizing membrane proteins on biosensor surfaces,109 as well as the need to maintain their active conformation in a lipid-like environment, complicates drug screening for membrane proteins. To address this issue, Cao and their research team utilized a method combining SPR with styrene-maleic acid (SMA) polymers, extracting P-gp from MCF-7/ADR (Adriamycin-resistant human breast cancer cell line) cells to form SMA lipid particles (SMALPs). This established a highly specific and stable screening system for P-gp inhibitors, successfully identifying 9 inhibitors with binding activity to P-gp from natural products.60 Similarly, Salipro nanoparticle system was utilized with saponin proteins to stabilize PANX1 protein extracted from crude cell microsomes in a quasi-natural lipid environment. Using SPR, the binding characteristics of this membrane protein with benzoylbenzoyl ATP (bzATP) and spironolactone were studied, determining the stabilization method of this membrane protein. The application of SPR technology in membrane protein drug research can accelerate the screening and study of membrane protein-targeted drugs in the future. Furthermore, in the treatment of malignant B-cell tumors, CD20 is considered an effective target.110 However, the difficulty in obtaining its recombinant or natural full-length protein has limited the development of anti-CD20 drugs. Compared to traditional lipid-based membrane protein stabilization techniques, extracellular vesicles (EVs) offer a new solution as a membrane protein display platform. The phospholipid bilayer of EVs provides an ideal microenvironment for the natural presentation of membrane proteins like CD20, helping to maintain their correct conformation and biological activity. Utilizing SPR technology, CD20-containing EVs can be indirectly captured on an SA chip, and specific anti-CD20 drugs, such as rituximab and obinutuzumab, can be injected to assess the specificity and targeting of the drugs. This method, integrating SPR technology, not only achieves accurate detection of CD20 and evaluation of drug targeting but also highlights the important role and broad application prospects of SPR technology in the biomedical field.62 Consequently, SPR can stably capture membrane proteins and maintain their activity, making it particularly suitable for studying membrane proteins like P-glycoprotein that are associated with multidrug resistance, thus accelerating the drug screening and development process. Additionally, this technology can be used to investigate the interactions and functions of other key membrane proteins, further advancing drug discovery and development.
In the development of detection tools, SPR technology has shown its significant role in the biomedical field. SPR technology finds extensive and profound applications in the development of detection tools. By utilizing the plasmon resonance phenomenon on metal surfaces, SPR enables real-time, label-free monitoring of biomolecular interactions. Annexin V protein, as a sensitive and specific probe, is widely used to mark cell apoptosis, especially due to its high affinity for phosphatidylserine (PS) exposure. Research was conducted using SPR technology to explore the application of sfGFP-ANXV (recombinant human annexin V produced as a fusion with a highly fluorescent superfolder derivative of the green fluorescent protein) fusion protein in the quantitative detection of liposomes or exosomes, thereby achieving rapid detection of cell apoptosis. By immobilizing liposomes on the chip surface and injecting three types of proteins: sfGFP-ANXV, ANXV, and sfGFP, the results showed that only sfGFP-ANXV had the ability to bind with immobilized liposomes.52 In cell biology, the translocation of membrane proteins to the plasma membrane (PM) is a key process involving various cellular functions, such as G protein-coupled receptor signaling, vesicular transport, and Ca2+ entry. To precisely control the membrane binding of target proteins in both spatial and temporal dimensions, researchers designed an optogenetic tool named OptoPB, which can selectively bind to the plasma membrane under blue light illumination and automatically dissociate back to the cytoplasm in the dark. Using SPR technology, researchers captured OptoPB on POPC liposomes containing 6% PI(4,5)P2 and found significant differences in the binding and dissociation constants of OptoPB under light and dark conditions. This not only confirmed the photosensitivity of OptoPB but also, through mutational analysis of different residues of OptoPB, further revealed the molecular mechanism of its binding to the plasma membrane.53 Two highly specific techniques were employed, namely sortase tagging and click chemistry, to graft AGuIX (activation and guidance of irradiation X) nanoparticles with two different targeting VHHs (Programmed Death-Ligand 1 (PD-L1, A12 VHH) and Cluster of Differentiation 47 (CD47, A4 VHH)), aiming for combined targeted immunotherapy. During this process, the efficacy of the two conjugation techniques was assessed through SPR experiments. The results showed that both bioconjugation methods produced AGuIX@VHH with strong binding affinity, verifying that they retained the functionality of the VHHs without affecting their binding capacity. Notably, the click chemistry method demonstrated a higher VHH conversion rate with the use of lower-cost reagents. Therefore, it is considered a favorable and promising approach for diverse applications in targeted therapeutic interventions and imaging.78 Therefore, the aforementioned studies highlight that combining SPR with lipid nanoparticles holds promising potential to robustly support the development of diagnostic reagents.52 SPR’s applications in exploring cellular processes provide valuable molecular-level insights for the development and application of optogenetic tools.53 Furthermore, the application of SPR technology provides crucial information for evaluating the effectiveness of two conjugation techniques, enzyme tagging and click chemistry, offering a reliable method to assess drug development tools.78 Therefore, SPR technology not only plays a crucial role in basic research but also drives the development and application of efficient and precise detection tools across various fields.
Future Prospects of SPR for Nanoparticle Application
Although SPR has made significant strides in the biological and chemical fields, many challenges remain to be addressed. Moreover, with continuous updates and advancements in SPR technology, its application to nanoparticle studies will significantly broaden the scope of research and development in nanotechnology.
SPR Enhanced Surface Modification and Detection Sensitization
SPR was employed to study the surface modification of nanoparticles, such as coating with polymers or ligands, to improve biocompatibility and reduce toxicity.111 By analyzing how these modifications affect the binding properties and interactions with target molecules, researchers can fine-tune the design of nanoparticles for specific applications. On the other hand, exploring the design and preparation of nanostructures can enhance SPR signals and improve detection sensitivity.112 Moreover, combining surface-enhanced Raman scattering (SERS) and localized surface plasmon resonance (LSPR) technologies will achieve more sensitive detection.113,114
Multimodal and Multiscale SPR
The focus on multimodal and multiscale SPR is expected to intensify.115 Multimodal and multiscale SPR refers to using SPR technology in different modes and at various scales for biomedical research and applications. Multimodal SPR involves employing SPR in combination with other techniques or modalities, such as surface-enhanced Raman scattering (SERS) or fluorescence, to enhance sensitivity or provide complementary information about molecular interactions. On the other hand, multiscale SPR involves utilizing SPR technology across different scales, ranging from microscale to nanoscale, to investigate interactions with varying levels of detail.116 Multimodal and multiscale SPR enable researchers to conduct thorough and comprehensive sample analyses across nano and micro scales, accommodating diverse types and sizes of samples. By exploring interactions among molecules, nanoparticles, and cells, this approach offers insights into intricate biological processes, thereby advancing the development of advanced biomedical diagnostics and therapies. Moreover, integration of SPR with ultra-performance liquid chromatography-mass spectrometry (UPLC-MS/MS), optical technologies, imaging systems, and electrochemistry will provide more comprehensive information.114,117–119
High-Throughput SPR Technology
High-throughput SPR refers to the application of SPR technology in a manner that allows for rapid and automated screening of interactions between nanoparticles and biomolecules. This approach facilitates the analysis of numerous samples simultaneously, enabling efficient assessment of binding kinetics, affinities, and specificities across a wide range of conditions. High-throughput SPR combines speed, automation, quantitative analysis, and versatility, making it a powerful tool for accelerating drug discovery, biomarker identification, and optimization of therapeutic candidates in biomedical research and pharmaceutical development.120,121 To meet the research and application needs in the biomedical field, the development of new technologies and devices based on high-throughput SPR technology will receive increased attention.
Development of SPR in Diagnostics Applications
SPR has already found numerous applications in clinical diagnostics and is expected to play an even more comprehensive role in disease diagnosis in future research. SPR offers label-free and real-time detection of biomolecular interactions, making it possible to detect minute changes in analyte concentrations. This capability is particularly suitable for identifying disease-related biomarkers, promising greater utility in early and sensitive detection.122 Additionally, SPR can distinguish between specific and non-specific binding events, enhancing the specificity of diagnostic assays for accurate disease diagnosis and monitoring.123 Multiplexed detection allows for the simultaneous analysis of multiple analytes within a single sample, making it ideal for screening and diagnosing complex diseases involving multiple biomarkers.124 By measuring changes in biomarker levels or drug-target interactions, SPR helps in assessing treatment outcomes and adjusting therapeutic strategies as needed. In the development of SPR instrumentation, portable and miniaturized devices have been created for point-of-care testing, enabling rapid diagnostic results directly at the patient’s bedside or in resource-limited settings.125 Furthermore, the integration of SPR with biosensors and microfluidic systems enhances its diagnostic applications.126,127 Biosensors coupled with SPR allow for real-time monitoring of biomolecular interactions in complex biological samples, further expanding its diagnostic potential. Consequently, the continuous advancement of SPR technology in diagnostics is improving its sensitivity, specificity, multiplexing capabilities, and suitability for point-of-care applications.105,128 These advancements are crucial for better disease detection, monitoring, and management, ultimately contributing to improved global healthcare outcomes. The continuous development and innovation of SPR in the diagnostic field will also bring new scenarios and opportunities for the broader research and application of nanoparticles in biomedicine field.
In summary, SPR technology is widely used for real-time, label-free detection of interactions between nanoparticles and biomolecules, which is crucial for accurately measuring binding affinities and kinetic parameters. SPR can assess the properties of nanoparticles and leverage them for signal amplification, facilitating the detection of low-concentration or low-abundance targets. Therefore, SPR technology is extensively applied in the biomedical use of nanoparticles, providing detailed insights into nanoparticle-biomolecule interactions and driving significant technological advancements in fields such as nanomaterial preparation, drug development, and clinical diagnostics. The further development and application of SPR technology will enhance its prospects in future nanomedicine research and beyond.
Conclusion
SPR, as an efficient tool for precise detection of molecular interactions, has demonstrated its unique value in the characterization of nanoparticles and the development of biologics. Currently, in the preparation and application research of biomedical nanoparticles, SPR technology is utilized for performance detection of nanoparticles, drug development and screening, and studying the interactions and mechanisms between nanoparticles and biomolecules. It plays a critical role in in-depth analysis of nanoparticle binding properties, screening and optimizing nanoparticle formulations, and detecting trace biomarkers, leading to significant research advancements and numerous commercial products based on SPR technology. However, challenges remain in the future research of nanoparticle performance characterization and applications. These challenges include regulating surface properties of nanoparticles to maintain stability in vivo, optimizing targeting precision and delivery efficiency, achieving large-scale production and batch consistency, and addressing long-term effects and potential risks in vivo. These issues involve the interactions between nanoparticles and biomolecules within the body. Therefore, resolving these problems largely depends on the continuous development and innovation of SPR technology. Due to its immense potential in detecting molecular interactions, SPR is expected to play an increasingly important role in the development of nanoparticle materials and biomedical applications. With ongoing technological innovation and application expansion, SPR is anticipated to open up broader application prospects in biomedical research and materials science.
SPR technology holds promising commercialization perspectives in biomedical fields due to its versatile applications and significant impact on research, diagnostics, and drug development. As SPR enables real-time, label-free detection of biomolecular interactions with high sensitivity and specificity, its adoption in pharmaceutical and biotechnology industries is expanding. In drug discovery, SPR facilitates rapid screening of drug candidates by measuring binding kinetics and affinity constants, thereby accelerating lead optimization and reducing development costs. This capability is particularly beneficial for evaluating target engagement, studying molecular mechanisms, and predicting drug efficacy. SPR is also pivotal in biomedical research, where it aids in studying protein-protein interactions, DNA-protein interactions, and antigen-antibody binding dynamics. This allows researchers to unravel disease mechanisms, identify biomarkers, and validate therapeutic targets. Furthermore, SPR technology plays a crucial role in diagnostic applications, including the detection of biomarkers for early disease diagnosis and monitoring therapeutic response. Its ability to provide quantitative and qualitative analysis of molecular interactions enhances diagnostic accuracy and improves patient outcomes. Commercially, SPR platforms are being integrated into automated systems for high-throughput screening and multiplexed assays, catering to the demand for faster and more efficient biomedical research tools. As the technology evolves, advancements in miniaturization and sensor design are enhancing portability and usability, expanding its potential use in point-of-care diagnostics and personalized medicine. SPR technology has evolved beyond traditional prism-based SPR to include grating-coupled SPR and waveguide-coupled SPR. Additionally, high-throughput SPR imaging (SPRi) and techniques based on the surface plasmon resonance of metal nanoparticles, such as localized surface plasmon resonance (LSPR) and surface-enhanced Raman scattering (SERS), have been developed to meet various demands. Overall, SPR technology’s capability to provide precise and real-time analysis of biomolecular interactions positions it as a valuable tool in advancing biomedical research, clinical diagnostics, and pharmaceutical development, driving its commercialization across diverse applications in the biomedical sector.
Abbreviations
SPR, surface plasmon resonance; GST, glutathione-S-transferase; EDTA, ethylenediaminetetraacetic acid; RU, resonance units; Aβ, amyloid-beta; AD, Alzheimer’s disease; PA, phosphatidic acid; CL, cardiolipin; PEGylated, polyethylene glycolated; BsmAb, monoclonal bispecific antibodies; CEA, carcino-embryonic; DTPA-In, diethylenetriaminepentaacetic acid-indium; DSPE, 1,2-Distearoyl-sn-glycero-3-phosphorylethanolamine; NPs, Nanoparticles; MIP NPs, molecularly imprinted polymer nanoparticles; SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2; COVID-19, Coronavirus Disease 2019; PD1, Programmed death-1; PD1-Imi Exo, PD1 gene engineered exosome enveloping imiquimod; PDL1, Programmed death-ligand 1; SNPs, self-assembling nanoparticles; DHD-glu-gln, dihexadecyl-glutamate-glutamine; DHD-glu-asn, dihexadecyl-glutamate-asparagine; DHD-glu-glu, dihexadecyl-glutamate-glutamic acid; PEGLip, polyethylene glycol-modified liposomes; ABX, an albumin-bound paclitaxel scaffold, CD20, B-lymphocyte antigen CD20; CDR H3, the third complementarity-determining regions of the heavy chain; ECH-Zn, echinacoside-zinc; HA-PEI, hyaluronic acid/poly(ethylenimine); PPZn, HA-PEI-coated ECH-Zn; RAGE, advanced glycation end-products receptor; MDM2, mouse double minute 2; STAT2, Signal transducer and activator of transcription 2; Cry4Ba-DIII, C-terminal domain (DIII) of the Cry4Ba mosquito-active toxin; BSA, bovine serum albumin; RBD, receptor-binding domain; aSA3, aSA3 nanobody; aRBD-2, aRBD-2 nanobody; CD40, cluster of differentiation 40; CD40L, CD40 ligand; MTX, methotrexate; B16, mouse melanoma cell line; MCF-7, human breast cancer cell line; RGD, Arg-Gly-Asp peptides, NRG1, neuregulin 1; Hu, anti-IGG-Fc; SARMs, Selective Androgen Receptor Modulators; MDR, multidrug resistance; P-gp, P-glycoprotein; SMA, styrene-maleic acid; SMALPs, SMA lipid particles; EVs, extracellular vesicles; B16, mouse melanoma cell line; MCF-7, human breast cancer cell line; MCF-7/ADR, Adriamycin-resistant human breast cancer cell line; bzATP, benzoylbenzoyl ATP; PS, phosphatidylserine; sfGFP, superfolder green fluorescent protein; ANXV, recombinant human annexin V; sfGFP-ANXV, recombinant human annexin V fused with a sfGFP; PM, plasma membrane; OptoPB, an optogenetic tool engineered by fusion of the phosphoinositide (PI)-binding polybasic domain of Rit1 (Rit-PB) to a photoreactive light-oxygen-voltage (LOV) domain; PI(4,5)P2, phosphatidylinositol 4,5-bisphosphate; POPC, 1-palmitoyl-2-oleoyl -glycero-3-phosphocholine; EBC, Exhaled Breath Condensate; QD-AB, InP/ZnS quantum dots conjugated antibodies; AGuIX, activation and guidance of irradiation X; VHH, heavy domain of heavy chain; UPLC-MS/MS, ultra-performance liquid chromatography-mass spectrometry; LSPR, localized surface plasmon resonance.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Petros RA, DeSimone JM. Strategies in the design of nanoparticles for therapeutic applications. Nat Rev Drug Discov. 2010;9(8):615–627. doi: 10.1038/nrd2591 [DOI] [PubMed] [Google Scholar]
- 2.Desfrancois C, Auzely R, Texier I. Lipid nanoparticles and their hydrogel composites for drug delivery: a review. Pharmaceuticals. 2018;11(4):118. doi: 10.3390/ph11040118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rohovie MJ, Nagasawa M, Swartz JR. Virus-like particles: next-generation nanoparticles for targeted therapeutic delivery. Bioeng Transl Med. 2017;2(1):43–57. doi: 10.1002/btm2.10049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Padmanabhan P, Kumar A, Kumar S, Chaudhary RK, Gulyas B. Nanoparticles in practice for molecular-imaging applications: an overview. Acta Biomater. 2016;41:1–16. doi: 10.1016/j.actbio.2016.06.003 [DOI] [PubMed] [Google Scholar]
- 5.Xu H, Cui Y, Tian Y, et al. Nanoparticle-based drug delivery systems for enhancing bone regeneration. ACS Biomater Sci Eng. 2024;10(3):1302–1322. doi: 10.1021/acsbiomaterials.3c01643 [DOI] [PubMed] [Google Scholar]
- 6.Sanchez-Gaytan BL, Fay F, Lobatto ME, et al. HDL-mimetic PLGA nanoparticle to target atherosclerosis plaque macrophages. Bioconjugate Chem. 2015;26(3):443–451. doi: 10.1021/bc500517k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pinto S, Hosseini M, Buckley ST, et al. Nanoparticles targeting the intestinal Fc receptor enhance intestinal cellular trafficking of semaglutide. J Control Release. 2024;366:621–636. doi: 10.1016/j.jconrel.2024.01.015 [DOI] [PubMed] [Google Scholar]
- 8.Qi Y, Kan Y, Li Z. High-resolution imaging of 3D stray-field components with a Fe3O4 nanoparticle sensor. Nanoscale. 2024;16(10):5164–5168. doi: 10.1039/D3NR05437C [DOI] [PubMed] [Google Scholar]
- 9.Luo L, Zhou H, Wang S, et al. The application of nanoparticle-based imaging and phototherapy for female reproductive organs diseases. Small. 2023. doi: 10.1002/smll.202207694 [DOI] [PubMed] [Google Scholar]
- 10.Klug J. Ku autoantigen is a potential major cause of nonspecific bands in electrophoretic mobility shift assays. Biotechniques. 1997;22(2):212–214, 216. doi: 10.2144/97222bm02 [DOI] [PubMed] [Google Scholar]
- 11.Aydin S. A short history, principles, and types of ELISA, and our laboratory experience with peptide/protein analyses using ELISA. Peptides. 2015;72:4–15. doi: 10.1016/j.peptides.2015.04.012 [DOI] [PubMed] [Google Scholar]
- 12.Jerabek-Willemsen M, André T, Wanner R, et al. MicroScale Thermophoresis: interaction analysis and beyond. J Mol Struct. 2014;1077:101–113. doi: 10.1016/j.molstruc.2014.03.009 [DOI] [Google Scholar]
- 13.Thillaivinayagalingam P, Gommeaux J, McLoughlin M, Collins D, Newcombe AR. Biopharmaceutical production: applications of surface plasmon resonance biosensors. J Chromatogr B Analyt Technol Biomed Life Sci. 2010;878(2):149–153. doi: 10.1016/j.jchromb.2009.08.040 [DOI] [PubMed] [Google Scholar]
- 14.Wulandari C, Septiani N, Gumilar G, et al. Surface plasmon resonance biosensor chips integrated with MoS-MoO hybrid microflowers for rapid CFP-10 tuberculosis detection. J Mat Chem B. 2023;11(48):11588–11599. doi: 10.1039/d3tb01327h [DOI] [PubMed] [Google Scholar]
- 15.Olaru A, Gheorghiu M, David S, Polonschii C, Gheorghiu E. Quality assessment of SPR sensor chips; case study on L1 chips. Biosens Bioelectron. 2013;45:77–81. doi: 10.1016/j.bios.2013.01.045 [DOI] [PubMed] [Google Scholar]
- 16.Arcadio F, Soares S, Nedoma J, et al. POF-based biosensors for cortisol detection in seawater as a tool for aquaculture systems. Sci Rep. 2024;14(1):13117. doi: 10.1038/s41598-024-63870-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei Y, Shi C, Zhang Y, et al. Spiral cone fiber SPR sensor for detecting ginsenoside Rg1. Opt Express. 2024;32(8):13783–13796. doi: 10.1364/OE.519188 [DOI] [PubMed] [Google Scholar]
- 18.Chen Y, Zhang H, Li R, et al. Novel multifunctional meta-surface plasmon resonance chip microplate for high-throughput molecular screening. Adv Healthc Mater. 2024:e2401097. doi: 10.1002/adhm.202401097 [DOI] [PubMed] [Google Scholar]
- 19.Chang S, Liu L, Mu C, et al. An Ultrasensitive SPR biosensor for RNA detection based on robust GeP nanosheets. J Colloid Interface Sci. 2023;651:938–947. doi: 10.1016/j.jcis.2023.08.064 [DOI] [PubMed] [Google Scholar]
- 20.Lei Z, Guo B. 2D material-based optical biosensor: status and prospect. Adv Sci. 2022;9(4):e2102924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Cao J, Zhang L, Liu Y, Liu Z, Chen H. 2D MOF-enhanced SPR detector based on tunable supramolecular probes for direct and sensitive detection of DOX in serum. Mikrochimica acta. 2024;191(3):154. doi: 10.1007/s00604-024-06226-2 [DOI] [PubMed] [Google Scholar]
- 22.Li J, Liu X, Sun H, et al. Optical fiber sensing probe for detecting a carcinoembryonic antigen using a composite sensitive film of PAN nanofiber membrane and gold nanomembrane. Opt Express. 2024;32(11):20024. doi: 10.1364/OE.523513 [DOI] [PubMed] [Google Scholar]
- 23.Qiu Q, Xu Y. Rapid and sensitive detection by combining electric field effects and surface plasmon resonance: a theoretical study. Micromachines. 2024;15(5):653. doi: 10.3390/mi15050653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaurav Pal S, Neha SJP. Smartphone-based surface plasmon resonance sensors: a review. Plasmonics. 2022;17(5):1869–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.S A, M J, C M, Maillart E. High performance multi-spectral interrogation for surface plasmon resonance imaging sensors. Biosens Bioelectron. 2013;54:175–180. [DOI] [PubMed] [Google Scholar]
- 26.Soocheol K, Jin Hwa R, Hoesung Y, et al. Spectrometer-based wavelength interrogation SPR imaging via Hadamard transform. Optics Letters. 2023;48(4):992–995. [DOI] [PubMed] [Google Scholar]
- 27.Dongping W, Fong-Chuen L, Hengji C, et al. Real-time multi-channel SPR sensing based on DMD-enabled angular interrogation. Optics Express. 2018;26(19):24627–24636. [DOI] [PubMed] [Google Scholar]
- 28.Yafeng H, Lulu Z, Hao Z, et al. Development of a portable SPR sensor for nucleic acid detection. Micromachines. 2020;11(5):526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jayeta B, Sudip M, Manik P. Polarization-multiplexed incoherent broadband surface plasmon resonance: a new analytical strategy for plasmonic sensing. Anal Chem. 2022;94(18):6689–6694. [DOI] [PubMed] [Google Scholar]
- 30.William OFC, M-SJM JR. Surface plasmon resonances in Sierpinski-like photonic crystal fibers: polarization filters and sensing applications. Molecules. 2020;25(20):4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nipun V, Marwan JA, Anand MS, Aabha B, Ibrahim AJB. Real-time ellipsometric surface plasmon resonance sensor using polarization camera may provide the ultimate detection limit. Biosensors. 2023;13(2):173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yinling X, Yu G, Jun G, Qian C, Juan X, Feimeng Z. Real-time detection of Cu2+ sequestration and release by immobilized apo-metallothioneins using SECM combined with SPR. Biosens Bioelectron. 2008;24(3):369–375. [DOI] [PubMed] [Google Scholar]
- 33.Gobbi M, Re F, Canovi M, et al. Lipid-based nanoparticles with high binding affinity for amyloid-β1-42 peptide. Biomaterials. 2010;31(25):6519–6529. doi: 10.1016/j.biomaterials.2010.04.044 [DOI] [PubMed] [Google Scholar]
- 34.Crielaard BJ, Yousefi A, Schillemans JP, et al. An in vitro assay based on surface plasmon resonance to predict the in vivo circulation kinetics of liposomes. J Control Release. 2011;156(3):307–314. doi: 10.1016/j.jconrel.2011.07.023 [DOI] [PubMed] [Google Scholar]
- 35.Seong H, Choi WM, Kim JC, Thompson DH, Park K. Preparation of liposomes with glucose binding sites: liposomes containing di-branched amino acid derivatives. Biomaterials. 2003;24(24):4487–4493. doi: 10.1016/S0142-9612(03)00352-1 [DOI] [PubMed] [Google Scholar]
- 36.Shi K, Long Y, Xu C, et al. Liposomes combined an integrin alphavbeta3-specific vector with pH-responsible cell-penetrating property for highly effective antiglioma therapy through the blood-brain barrier. ACS Appl Mater Interfaces. 2015;7(38):21442–21454. doi: 10.1021/acsami.5b06429 [DOI] [PubMed] [Google Scholar]
- 37.Terada T, Mizobata M, Kawakami S, Yamashita F, Hashida M. Optimization of tumor-selective targeting by basic fibroblast growth factor-binding peptide grafted PEGylated liposomes. J Control Release. 2007;119(3):262–270. doi: 10.1016/j.jconrel.2007.01.018 [DOI] [PubMed] [Google Scholar]
- 38.Al-Ahmady ZS, Chaloin O, Kostarelos K. Monoclonal antibody-targeted, temperature-sensitive liposomes: in vivo tumor chemotherapeutics in combination with mild hyperthermia. J Control Release. 2014;196:332–343. doi: 10.1016/j.jconrel.2014.10.013 [DOI] [PubMed] [Google Scholar]
- 39.Huang ZR, Tipparaju SK, Kirpotin DB, et al. Formulation optimization of an ephrin A2 targeted immunoliposome encapsulating reversibly modified taxane prodrugs. J Control Release. 2019;310:47–57. doi: 10.1016/j.jconrel.2019.08.006 [DOI] [PubMed] [Google Scholar]
- 40.Yatuv R, Carmel-Goren L, Dayan I, Robinson M, Baru M. Binding of proteins to PEGylated liposomes and improvement of G-CSF efficacy in mobilization of hematopoietic stem cells. J Control Release. 2009;135(1):44–50. doi: 10.1016/j.jconrel.2008.12.004 [DOI] [PubMed] [Google Scholar]
- 41.Ding Q, Si X, Liu D, et al. Targeting and liposomal drug delivery to CD40L expressing T cells for treatment of autoimmune diseases. J Control Release. 2015;207:86–92. doi: 10.1016/j.jconrel.2015.03.035 [DOI] [PubMed] [Google Scholar]
- 42.Guo Z, He B, Jin H, et al. Targeting efficiency of RGD-modified nanocarriers with different ligand intervals in response to integrin alphavbeta3 clustering. Biomaterials. 2014;35(23):6106–6117. doi: 10.1016/j.biomaterials.2014.04.031 [DOI] [PubMed] [Google Scholar]
- 43.Sugano M, Morisaki H, Negishi Y, et al. Potential effect of cationic liposomes on interactions with oral bacterial cells and biofilms. J Liposome Res. 2016;26(2):156–162. doi: 10.3109/08982104.2015.1063648 [DOI] [PubMed] [Google Scholar]
- 44.Rauscher A, Frindel M, Maurel C, et al. Influence of pegylation and hapten location at the surface of radiolabelled liposomes on tumour immunotargeting using bispecific antibody. Nucl Med Biol. 2014;41(Suppl):e66–e74. doi: 10.1016/j.nucmedbio.2013.12.012 [DOI] [PubMed] [Google Scholar]
- 45.Satoh A, Miwa HE, Kojima K, Hirabayashi J, Matsumoto I. Ligand-binding properties of annexin from Caenorhabditis elegans (annexin XVI, Nex-1). J Biochem. 2000;128(3):377–381. doi: 10.1093/oxfordjournals.jbchem.a022764 [DOI] [PubMed] [Google Scholar]
- 46.Gil C, Dorca-Arevalo J, Blasi J. Calcium enhances binding of Clostridium perfringens epsilon toxin to sulfatide. Biochim Biophys Acta Biomembr. 2019;1861(1):161–169. doi: 10.1016/j.bbamem.2018.08.003 [DOI] [PubMed] [Google Scholar]
- 47.Lombardi D, Cuenoud B, Wunderli-Allenspach H, Kramer SD. Interaction kinetics of salmeterol with egg phosphatidylcholine liposomes by surface plasmon resonance. Anal Biochem. 2009;385(2):215–223. doi: 10.1016/j.ab.2008.11.011 [DOI] [PubMed] [Google Scholar]
- 48.Budragchaa D, Shiming B, Kanamoto T, Nakashima H, Miyazaki K, Yoshida T. Interaction between sulfated 3-O-octadecyl-alpha-(1-->6)-d-glucan and liposomes analyzed by surface plasmon resonance. Carbohydr Polym. 2020;239:116022. doi: 10.1016/j.carbpol.2020.116022 [DOI] [PubMed] [Google Scholar]
- 49.Mingxue B, Chaolumen B, Asai D, Takemura H, Miyazaki K, Yoshida T. Role of a long-chain alkyl group in sulfated alkyl oligosaccharides with high anti-HIV activity revealed by SPR and DLS. Carbohydr Polym. 2020;245:116518. doi: 10.1016/j.carbpol.2020.116518 [DOI] [PubMed] [Google Scholar]
- 50.Shibata H, Saito H, Yomota C, Kawanishi T, Okuda H. Alterations in the detergent-induced membrane permeability and solubilization of saturated phosphatidylcholine/cholesterol liposomes: effects of poly(ethylene glycol)-conjugated lipid. Chem Pharm Bull. 2012;60(9):1105–1111. doi: 10.1248/cpb.c12-00153 [DOI] [PubMed] [Google Scholar]
- 51.Akkilic N, Liljeblad M, Blaho S, Holtta M, Hook F, Geschwindner S. Avidity-based affinity enhancement using nanoliposome-amplified SPR sensing enables low picomolar detection of biologically active neuregulin 1. ACS Sens. 2019;4(12):3166–3174. doi: 10.1021/acssensors.9b01392 [DOI] [PubMed] [Google Scholar]
- 52.Abbady AQ, Twair A, Ali B, Murad H. Characterization of annexin V fusion with the superfolder GFP in liposomes binding and apoptosis detection. Front Physiol. 2017;8:317. doi: 10.3389/fphys.2017.00317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li L, He L, Wu B, et al. Structural determinants for light-dependent membrane binding of a photoswitchable polybasic domain. ACS Synth Biol. 2021;10(3):542–551. doi: 10.1021/acssynbio.0c00571 [DOI] [PubMed] [Google Scholar]
- 54.Etzerodt A, Maniecki MB, Graversen JH, Moller HJ, Torchilin VP, Moestrup SK. Efficient intracellular drug-targeting of macrophages using stealth liposomes directed to the hemoglobin scavenger receptor CD163. J Control Release. 2012;160(1):72–80. doi: 10.1016/j.jconrel.2012.01.034 [DOI] [PubMed] [Google Scholar]
- 55.Rafique A, Etzerodt A, Graversen JH, Moestrup SK, Dagnaes-Hansen F, Moller HJ. Targeted lipid nanoparticle delivery of calcitriol to human monocyte-derived macrophages in vitro and in vivo: investigation of the anti-inflammatory effects of calcitriol. Int J Nanomed. 2019;14:2829–2846. doi: 10.2147/IJN.S192113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Okamoto A, Asai T, Kato H, et al. Antibody-modified lipid nanoparticles for selective delivery of siRNA to tumors expressing membrane-anchored form of HB-EGF. Biochem Biophys Res Commun. 2014;449(4):460–465. doi: 10.1016/j.bbrc.2014.05.043 [DOI] [PubMed] [Google Scholar]
- 57.Mizrahy S, Raz SR, Hasgaard M, et al. Hyaluronan-coated nanoparticles: the influence of the molecular weight on CD44-hyaluronan interactions and on the immune response. J Control Release. 2011;156(2):231–238. doi: 10.1016/j.jconrel.2011.06.031 [DOI] [PubMed] [Google Scholar]
- 58.Thammasittirong A, Imtong C, Sriwimol W, Sakdee S, Angsuthanasombat C. The C-terminal domain of the Bacillus thuringiensis Cry4Ba mosquito-specific toxin serves as a potential membrane anchor. Toxins. 2019;11(2):62. doi: 10.3390/toxins11020062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mourtas S, Canovi M, Zona C, et al. Curcumin-decorated nanoliposomes with very high affinity for amyloid-β1-42 peptide. Biomaterials. 2011;32(6):1635–1645. doi: 10.1016/j.biomaterials.2010.10.027 [DOI] [PubMed] [Google Scholar]
- 60.Cao Y, Fang J, Shi Y, et al. Screening potential P-glycoprotein inhibitors by combination of a detergent-free membrane protein extraction with surface plasmon resonance biosensor. Acta Pharm Sin B. 2022;12(7):3113–3123. doi: 10.1016/j.apsb.2022.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Drulyte I, Gutgsell AR, Lloris-Garcera P, et al. Direct cell extraction of membrane proteins for structure-function analysis. Sci Rep. 2023;13(1):1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang X, Phan MM, Sun Y, et al. Development of an SPR-based binding assay for characterization of anti-CD20 antibodies to CD20 expressed on extracellular vesicles. Anal Biochem. 2022;646:114635. doi: 10.1016/j.ab.2022.114635 [DOI] [PubMed] [Google Scholar]
- 63.Dobhal G, Datta A, Ayupova D, Teesdale-Spittle P, Goreham RV. Isolation, characterisation and detection of breath-derived extracellular vesicles. Sci Rep. 2020;10(1):17381. doi: 10.1038/s41598-020-73243-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li P, Xie Y, Wang J, et al. Gene engineered exosome reverses T cell exhaustion in cancer immunotherapy. Bioact Mater. 2024;34:466–481. doi: 10.1016/j.bioactmat.2024.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Obuobi S, Wang Y, Khara JS, Riegger A, Kuan SL, Ee PLR. Antimicrobial and anti-biofilm activities of surface engineered polycationic albumin nanoparticles with reduced hemolytic activity. Macromol Biosci. 2018;18(10):e1800196. doi: 10.1002/mabi.201800196 [DOI] [PubMed] [Google Scholar]
- 66.Henderson A, Sullivan MV, Hand RA, Turner NW. Detection of selective androgen receptor modulators (SARMs) in serum using a molecularly imprinted nanoparticle surface plasmon resonance sensor. J Mater Chem B. 2022;10(35):6792–6799. doi: 10.1039/D2TB00270A [DOI] [PubMed] [Google Scholar]
- 67.Hong S, Leroueil PR, Majoros IJ, Orr BG, Baker JR, Banaszak Holl MM. The binding avidity of a nanoparticle-based multivalent targeted drug delivery platform. Chem Biol. 2007;14(1):107–115. doi: 10.1016/j.chembiol.2006.11.015 [DOI] [PubMed] [Google Scholar]
- 68.Safaryan AHM, Smith AM, Bedwell TS, Piletska EV, Canfarotta F, Piletsky SA. Optimisation of the preservation conditions for molecularly imprinted polymer nanoparticles specific for trypsin. Nanoscale Adv. 2019;1(9):3709–3714. doi: 10.1039/C9NA00327D [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shrivastava S, Carmen JM, Lu Z, et al. SARS-CoV-2 spike-ferritin-nanoparticle adjuvanted with ALFQ induces long-lived plasma cells and cross-neutralizing antibodies. Npj Vaccines. 2023;8(1):43. doi: 10.1038/s41541-023-00638-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lu RM, Liang KH, Chiang HL, et al. Broadly neutralizing antibodies against Omicron variants of SARS-CoV-2 derived from mRNA-lipid nanoparticle-immunized mice. Heliyon. 2023;9(5):e15587. doi: 10.1016/j.heliyon.2023.e15587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li R, Chang Z, Liu H, et al. Double-layered N-S1 protein nanoparticle immunization elicits robust cellular immune and broad antibody responses against SARS-CoV-2. J Nanobiotechnol. 2024;22(1):44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang X, Wu S, Liu J, et al. A mosaic nanoparticle vaccine elicits potent mucosal immune response with significant cross-protection activity against multiple SARS-CoV-2 sublineages. Adv Sci. 2023;10(27):e2301034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhen JB, Zhao MH, Ge Y, et al. Construction, mechanism, and antibacterial resistance insight into polypeptide-based nanoparticles. Biomater Sci. 2019;7(10):4142–4152. doi: 10.1039/C9BM01050E [DOI] [PubMed] [Google Scholar]
- 74.Xu Y, Deng M, Zhang H, et al. Selection of affinity reagents to neutralize the hemolytic toxicity of melittin based on a self-assembled nanoparticle library. ACS Appl Mater Interfaces. 2020;12(14):16040–16049. doi: 10.1021/acsami.0c00303 [DOI] [PubMed] [Google Scholar]
- 75.Gabold B, Adams F, Brameyer S, et al. Transferrin-modified chitosan nanoparticles for targeted nose-to-brain delivery of proteins. Drug Deliv Transl Res. 2023;13(3):822–838. doi: 10.1007/s13346-022-01245-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nevala WK, Butterfield JT, Sutor SL, Knauer DJ, Markovic SN. Antibody-targeted paclitaxel loaded nanoparticles for the treatment of CD20(+) B-cell lymphoma. Sci Rep. 2017;7(1):45682. doi: 10.1038/srep45682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Butterfield JT, Kim H, Knauer DJ, Nevala WK, Markovic SN. Identification of a peptide-peptide binding motif in the coating of nab-paclitaxel nanoparticles with clinical antibodies: bevacizumab, rituximab, and trastuzumab. Sci Rep. 2017;7(1):14476. doi: 10.1038/s41598-017-15251-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carmès L, Bort G, Lux F, et al. AGuIX nanoparticle-nanobody bioconjugates to target immune checkpoint receptors. Nanoscale. 2024;16(5):2347–2360. doi: 10.1039/D3NR04777F [DOI] [PubMed] [Google Scholar]
- 79.Gao LY, Liu XY, Chen CJ, et al. Core-shell type lipid/rPAA-Chol polymer hybrid nanoparticles for in vivo siRNA delivery. Biomaterials. 2014;35(6):2066–2078. doi: 10.1016/j.biomaterials.2013.11.046 [DOI] [PubMed] [Google Scholar]
- 80.Ma H, Zhang X, Zeng W, et al. A bispecific nanobody dimer broadly neutralizes SARS-CoV-1 & 2 variants of concern and offers substantial protection against Omicron via low-dose intranasal administration. Cell Discov. 2022;8(1):132. doi: 10.1038/s41421-022-00497-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pan H, Ivashyna O, Sinha B, et al. Post-formulation peptide drug loading of nanostructures for metered control of NF-kappaB signaling. Biomaterials. 2011;32(1):231–238. doi: 10.1016/j.biomaterials.2010.08.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sun T, Zhang Y, Zhao F, Xia N, Liu L. Self-assembled biotin-phenylalanine nanoparticles for the signal amplification of surface plasmon resonance biosensors. Mikrochim Acta. 2020;187(8):473. doi: 10.1007/s00604-020-04461-x [DOI] [PubMed] [Google Scholar]
- 83.Han J, Sun Y, Wu T, et al. Echinacoside-Zinc nanomaterial inhibits skin glycation by suppressing the transcriptional activation of the receptor for advanced glycation end-products. ACS Nano. 2023;17(14):14123–14135. doi: 10.1021/acsnano.3c04726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Piletska EV, Piletsky SA. Size matters: influence of the size of nanoparticles on their interactions with ligands immobilized on the solid surface. Langmuir. 2010;26(6):3783–3785. doi: 10.1021/la904834y [DOI] [PubMed] [Google Scholar]
- 85.Belen SM, Sofia NT, Romina M, et al. Optimized surface plasmon resonance immunoassay for staphylococcal enterotoxin G detection using silica nanoparticles. Biochem Biophys Res Commun. 2021;558:168–174. doi: 10.1016/j.bbrc.2021.04.077 [DOI] [PubMed] [Google Scholar]
- 86.Findeis MA. The role of amyloid β peptide 42 in Alzheimer’s disease. Pharmacol Ther. 2007;116(2):266–286. doi: 10.1016/j.pharmthera.2007.06.006 [DOI] [PubMed] [Google Scholar]
- 87.Farshchi F, Dias-Lopes G, Castro-Côrtes LM, Alves CR, Souza-Silva F. Recent advances in surface plasmon resonance as a powerful approach for studying Leishmania spp. and Trypanosoma cruzi parasites. Talanta Open. 2023;8:100266. doi: 10.1016/j.talo.2023.100266 [DOI] [Google Scholar]
- 88.Mallick S, Fau - Choi JS, Choi JS. Liposomes: versatile and biocompatible nanovesicles for efficient biomolecules delivery. J Nanosci Nanotechnol. 2014;14(1):755–765. doi: 10.1166/jnn.2014.9080 [DOI] [PubMed] [Google Scholar]
- 89.Li P, Zhang M, Zou Y, et al. Interaction of heat shock protein 90 B1 (Hsp90B1) with liposome reveals its potential role in protection the integrity of lipid membranes. Int J Biol Macromol. 2018;106:1250–1257. doi: 10.1016/j.ijbiomac.2017.08.121 [DOI] [PubMed] [Google Scholar]
- 90.Man Z, Weizhen F, Ying W, et al. M335, a novel small-molecule STING agonist activates the immune response and exerts antitumor effects. Eur J Med Chem. 2023;264:116018. [DOI] [PubMed] [Google Scholar]
- 91.Rudson JH, Candida D, Leandro SM-D, et al. Plasmodium falciparum purine nucleoside phosphorylase as a model in the search for new inhibitors by high throughput screening. Int J Biol Macromol. 2020;165:1832–1841. [DOI] [PubMed] [Google Scholar]
- 92.Giulia C, Fabio MS, Yessica M, et al. Label-free detection of tobramycin in serum by transmission-localized surface plasmon resonance. Anal Chem. 2015;87(10):5278–5285. [DOI] [PubMed] [Google Scholar]
- 93.Chang L, Subbiah A, Haitham AB, et al. Live cell integrated surface plasmon resonance biosensing approach to mimic the regulation of angiogenic switch upon anti-cancer drug exposure. Anal Chem. 2014;86(15):7305–7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Preejith V, Binxing L, Kelly N, Paul SB. Surface plasmon resonance (SPR) studies on the interactions of carotenoids and their binding proteins. Arch Biochem Biophys. 2012;519(1):32–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nattanan J, Lueacha T, Duangnapa K, et al. A novel cyclic NP1 reveals obstruction of EGFR kinase activity and attenuation of EGFR-driven cell lines. J Cell Biochem. 2021;123(2):248–258. [DOI] [PubMed] [Google Scholar]
- 96.John TI, Susan LD, Lizamma A, et al. Third generation quinoline-3-carboxamide transcriptional disrupter of HDAC4, HIF-1α, and MEF-2 signaling for metastatic castration-resistant prostate cancer. Prostate. 2023;83(15):1470–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Walter H. A new strategy for improved secondary screening and lead optimization using high-resolution SPR characterization of compound-target interactions. J Mol Recognit. 2005;18(4):273–281. [DOI] [PubMed] [Google Scholar]
- 98.Ping L, Zesen C, Keyu M, et al. Discovery of taurocholic acid sodium hydrate as a novel repurposing drug for intervertebral disc degeneration by targeting MAPK3. Orthop Surg. 2023;16(1):183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vicki TT, David CH, Gregory JM, et al. Theoretical and experimental analysis of photon counting detector CT for proton stopping power prediction. Med Phys. 2018;45(11):5186–5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li G, Song Z, Ru Y, et al. Small-molecule nanoprodrug with high drug loading and EGFR, PI3K/AKT dual‐inhibiting properties for bladder cancer treatment. Exploration. 2023;3(5):20220141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tokuda T, Calero M, Fau - Matsubara E, et al. Lipidation of apolipoprotein E influences its isoform-specific interaction with Alzheimer’s amyloid beta peptides. Biochem J. 2000;348(2):359–365. doi: 10.1042/bj3480359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lim M, Fletcher NL, Saunus JM, et al. Targeted hyperbranched nanoparticles for delivery of doxorubicin in breast cancer brain metastasis. Mol Pharmaceut. 2023;20(12):6169–6183. doi: 10.1021/acs.molpharmaceut.3c00558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen C, YuWei Z, XinPeng N, et al. Directly targeting ASC by lonidamine alleviates inflammasome-driven diseases. J Neuroinflammation 2022;19(1):315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Suzanne OC, Yann-Vaï LB, Isaac MW, et al. Discovery and characterization of a cryptic secondary binding site in the molecular chaperone HSP70. Molecules. 2022;27(3):817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Singh AK, Anwar M, Pradhan R, Ashar MS, Rai N, Dey S. Surface plasmon resonance based-optical biosensor: emerging diagnostic tool for early detection of diseases. J Biophotonics. 2023;16(7):e202200380. doi: 10.1002/jbio.202200380 [DOI] [PubMed] [Google Scholar]
- 106.Deng N, Wickstrom L, Cieplak P, Lin C, Yang DJTjopc B. Resolving the ligand-binding specificity in c-MYC G-quadruplex DNA: absolute binding free energy calculations and SPR experiment. J Phys Chem. 2017;121(46):10484–10497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Martinez W, Arenas J, Mok L, et al. Bioelectronic measurement of target engagement to a membrane-bound transporter. SLAS Discovr. 2021;26(8):1004–1013. doi: 10.1177/24725552211013067 [DOI] [PubMed] [Google Scholar]
- 108.Abdiche YN, Myszka DG. Probing the mechanism of drug/lipid membrane interactions using Biacore. Anal Biochem. 2004;328(2):233–243. doi: 10.1016/j.ab.2004.01.018 [DOI] [PubMed] [Google Scholar]
- 109.Maynard JA, Lindquist NC, Sutherland JN, et al. Surface plasmon resonance for high‐throughput ligand screening of membrane‐bound proteins. Biotechnol J. 2009;4(11):1542–1558. doi: 10.1002/biot.200900195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rafiq S, Butchar JP, Cheney C, et al. Comparative assessment of clinically utilized CD20-directed antibodies in chronic lymphocytic leukemia cells reveals divergent NK cell, monocyte, and macrophage properties. J Immunol. 2013;190(6):2702–2711. doi: 10.4049/jimmunol.1202588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.He X, Hao T, Geng H, et al. Sensitization strategies of lateral flow immunochromatography for gold modified nanomaterials in biosensor development. Int J Nanomed. 2023;18:7847–7863. doi: 10.2147/IJN.S436379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bezuneh T, Fereja T, Kitte S, Li H, Jin YJT. Gold nanoparticle-based signal amplified electrochemiluminescence for biosensing applications. Talanta. 2022;248:123611. doi: 10.1016/j.talanta.2022.123611 [DOI] [PubMed] [Google Scholar]
- 113.Markina NE, Ustinov SN, Zakharevich AM, Markin AV. Copper nanoparticles for SERS-based determination of some cephalosporin antibiotics in spiked human urine. Anal Chim Acta. 2020;1138:9–17. doi: 10.1016/j.aca.2020.09.016 [DOI] [PubMed] [Google Scholar]
- 114.Li Q, Huo H, Wu Y, et al. Design and synthesis of SERS materials for in vivo molecular imaging and biosensing. Adv Sci. 2023;10(8):e2202051. doi: 10.1002/advs.202202051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cennamo N, Arcadio F, Seggio M, et al. Spoon-shaped polymer waveguides to excite multiple plasmonic phenomena: a multisensor based on antibody and molecularly imprinted nanoparticles to detect albumin concentrations over eight orders of magnitude. Biosens Bioelectron. 2022;217:114707. doi: 10.1016/j.bios.2022.114707 [DOI] [PubMed] [Google Scholar]
- 116.Durdagi S, Avsar T, Orhan M, et al. The neutralization effect of montelukast on SARS-CoV-2 is shown by multiscale in silico simulations and combined in vitro studies. Mol Ther. 2022;30(2):963–974. doi: 10.1016/j.ymthe.2021.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang HA-OX, Li XA-O, Zhou X, Zhang YA-O, Zhao YA-O. A lipase-conjugated carbon nanotube fiber-optic SPR sensor for sensitive and specific detection of tributyrin. Nanoscale. 2024;16(6):3113–3120. doi: 10.1039/D3NR05129C [DOI] [PubMed] [Google Scholar]
- 118.Liu D, Li Q, Luo J, Huang Q, Zhang Y. An SPRI beads-based DNA purification strategy for flexibility and cost-effectiveness. BMC Genomics. 2023;24(1):125. doi: 10.1186/s12864-023-09211-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ribeiro JA, Sales MGF, Pereira CM. Electrochemistry-assisted surface plasmon resonance biosensor for detection of CA 15-3. Anal Chem. 2021;93(22):7815–7824. doi: 10.1021/acs.analchem.0c05367 [DOI] [PubMed] [Google Scholar]
- 120.Martin HL, Turner AL, Higgins J, et al. Affimer-mediated locking of p21-activated kinase 5 in an intermediate activation state results in kinase inhibition. Cell Rep. 2023;42(10):113184. doi: 10.1016/j.celrep.2023.113184 [DOI] [PubMed] [Google Scholar]
- 121.Duan Y, Sun W, Li Y, et al. Spirohypertones A and B as potent antipsoriatics: tumor necrosis factor-α inhibitors with unprecedented chemical architectures. Acta Pharmaceutica Sinica B. 2024;14(6):2646–2656. doi: 10.1016/j.apsb.2024.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Choi J, Lee J, Son J, Choi JJS. Noble metal-assisted surface plasmon resonance immunosensors. Sensors. 2020;20(4):1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Choudhary S, Altintas ZJB. Development of a point-of-care SPR Sensor for the diagnosis of acute myocardial infarction. Biosensors. 2023;13(2):229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Khire T, Gao W, Bales B, et al. Rapid Minimum Inhibitory Concentration (MIC) analysis using lyophilized reagent beads in a novel multiphase, single-vessel assay. Antibiotics. 2023;12(11):1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mostufa S, Rezaei B, Ciannella S, et al. Advancements and perspectives in optical biosensors. ACS omega. 2024;9(23):24181–24202. doi: 10.1021/acsomega.4c01872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Khaksari S, Abnous K, Hadizadeh F, Ramezani M, Taghdisi S, Mousavi Shaegh SJT. Signal amplification strategies in biosensing of extracellular vesicles (EVs). Talanta. 2023;256:124244. doi: 10.1016/j.talanta.2022.124244 [DOI] [PubMed] [Google Scholar]
- 127.Qu J, Ordutowski H, Van Tricht C, et al. Point-of-care therapeutic drug monitoring of Adalimumab by integrating a FO-SPR biosensor in a self-powered microfluidic cartridge. Biosens Bioelectron. 2022;206:114125. doi: 10.1016/j.bios.2022.114125 [DOI] [PubMed] [Google Scholar]
- 128.Lee S, Dang H, Moon JI, et al. SERS-based microdevices for use as in vitro diagnostic biosensors. Chem Soc Rev. 2024;53(11):5394–5427. doi: 10.1039/d3cs01055d [DOI] [PubMed] [Google Scholar]