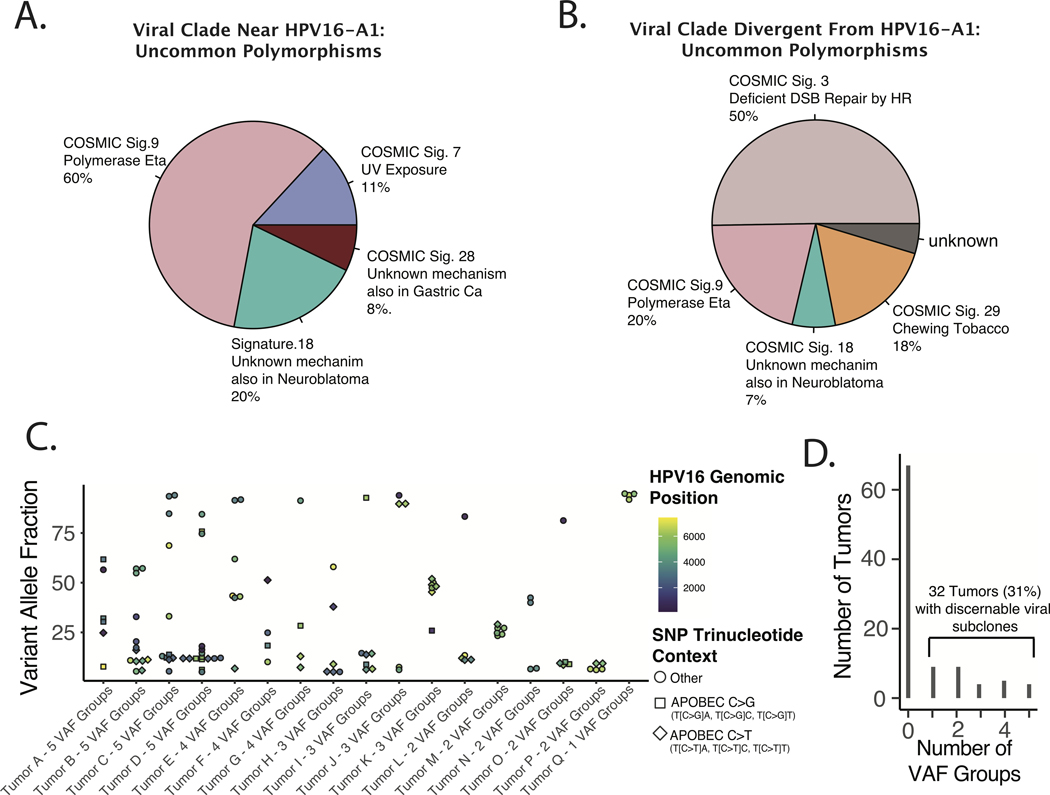
Figure 5. Origins of Environmental and Intra-tumoral Genomic Diversity of Oncogenic HPV16 in HNSCC.
A-B. Mutational Signature Analysis of Uncommon HPV16 Non-synonymous Clonal (Environmental) Polymorphisms. All non-synonymous polymorphisms identified in less than 25% of tumors were included. Non-negative matrix decomposition of all included SNPs was performed based on the COSMIC signatures V2, as implemented by the DeconstructSigs R package. A. Tumors in the viral clade near the HPV16-A1 reference sequence. B. Viral genomes distal to relative to HPV16-A1. C-E. Estimating HPV16 Sub-clonality with Binomial Mixture Clustering Analysis. C. Scatter plot of Sub-clone Defining Viral SNPs. VAF – variant allele frequency. D. Bar Plot of frequency of tumors by number of HPV16 sub-clonal populations. Populations defined by VAF Groups defined by binomial mixture clustering with BIC analysis as implemented in the R Canopy package.